Graphical Summarization of Continuous Variables Against a Response
summaryRc.RdsummaryRc is a continuous version of summary.formula
with method='response'. It uses the plsmo
function to compute the possibly stratified lowess
nonparametric regression estimates, and plots them along with the data
density, with selected quantiles of the overall distribution (over
strata) of each x shown as arrows on top of the graph. All the
x variables must be numeric and continuous or nearly continuous.
summaryRc(formula, data=NULL, subset=NULL,
na.action=NULL, fun = function(x) x,
na.rm = TRUE, ylab=NULL, ylim=NULL, xlim=NULL,
nloc=NULL, datadensity=NULL,
quant = c(0.05, 0.1, 0.25, 0.5, 0.75,
0.90, 0.95), quantloc=c('top','bottom'),
cex.quant=.6, srt.quant=0,
bpplot = c('none', 'top', 'top outside', 'top inside', 'bottom'),
height.bpplot=0.08,
trim=NULL, test = FALSE, vnames = c('labels', 'names'), ...)Arguments
- formula
An R formula with additive effects. The
formulamay contain one or more invocations of thestratifyfunction whose arguments are defined below. This causes the entire analysis to be stratified by cross-classifications of the combined list of stratification factors. This stratification will be reflected as separatelowesscurves.- data
name or number of a data frame. Default is the current frame.
- subset
a logical vector or integer vector of subscripts used to specify the subset of data to use in the analysis. The default is to use all observations in the data frame.
- na.action
function for handling missing data in the input data. The default is a function defined here called
na.retain, which keeps all observations for processing, with missing variables or not.- fun
function for transforming
lowessestimates. Default is the identity function.- na.rm
TRUE(the default) to excludeNAs before passing data tofunto compute statistics,FALSEotherwise.- ylab
y-axis label. Default is label attribute ofyvariable, or its name.- ylim
y-axis limits. By default each graph is scaled on its own.- xlim
a list with elements named as the variable names appearing on the
x-axis, with each element being a 2-vector specifying lower and upper limits. Any variable not appearing in the list will have its limits computed and possiblytrimmed.- nloc
location for sample size. Specify
nloc=FALSEto suppress, ornloc=list(x=,y=)wherex,yare relative coordinates in the data window. Default position is in the largest empty space.- datadensity
see
plsmo. Defaults toTRUEif there is astratifyvariable,FALSEotherwise.- quant
vector of quantiles to use for summarizing the marginal distribution of each
x. This must be numbers between 0 and 1 inclusive. UseNULLto omit quantiles.- quantloc
specify
quantloc='bottom'to place at the bottom of each plot rather than the default- cex.quant
character size for writing which quantiles are represented. Set to
0to suppress quantile labels.- srt.quant
angle for text for quantile labels
- bpplot
if not
'none'will draw extended box plot at location given bybpplot, and quantiles discussed above will be suppressed. Specifyingbpplot='top'is the same as specifyingbpplot='top inside'.- height.bpplot
height in inches of the horizontal extended box plot
- trim
The default is to plot from the 10th smallest to the 10th largest
xif the number of non-NAs exceeds 200, otherwise to use the entire range ofx. Specify another quantile to use other limits, e.g.,trim=0.01will use the first and last percentiles- test
Set to
TRUEto plot test statistics (not yet implemented).- vnames
By default, plots are usually labeled with variable labels (see the
labelandsas.getfunctions). To use the shorter variable names, specifyvnames="names".- ...
arguments passed to
plsmo
Value
no value is returned
See also
Examples
options(digits=3)
set.seed(177)
sex <- factor(sample(c("m","f"), 500, rep=TRUE))
age <- rnorm(500, 50, 5)
bp <- rnorm(500, 120, 7)
units(age) <- 'Years'; units(bp) <- 'mmHg'
label(bp) <- 'Systolic Blood Pressure'
L <- .5*(sex == 'm') + 0.1 * (age - 50)
y <- rbinom(500, 1, plogis(L))
par(mfrow=c(1,2))
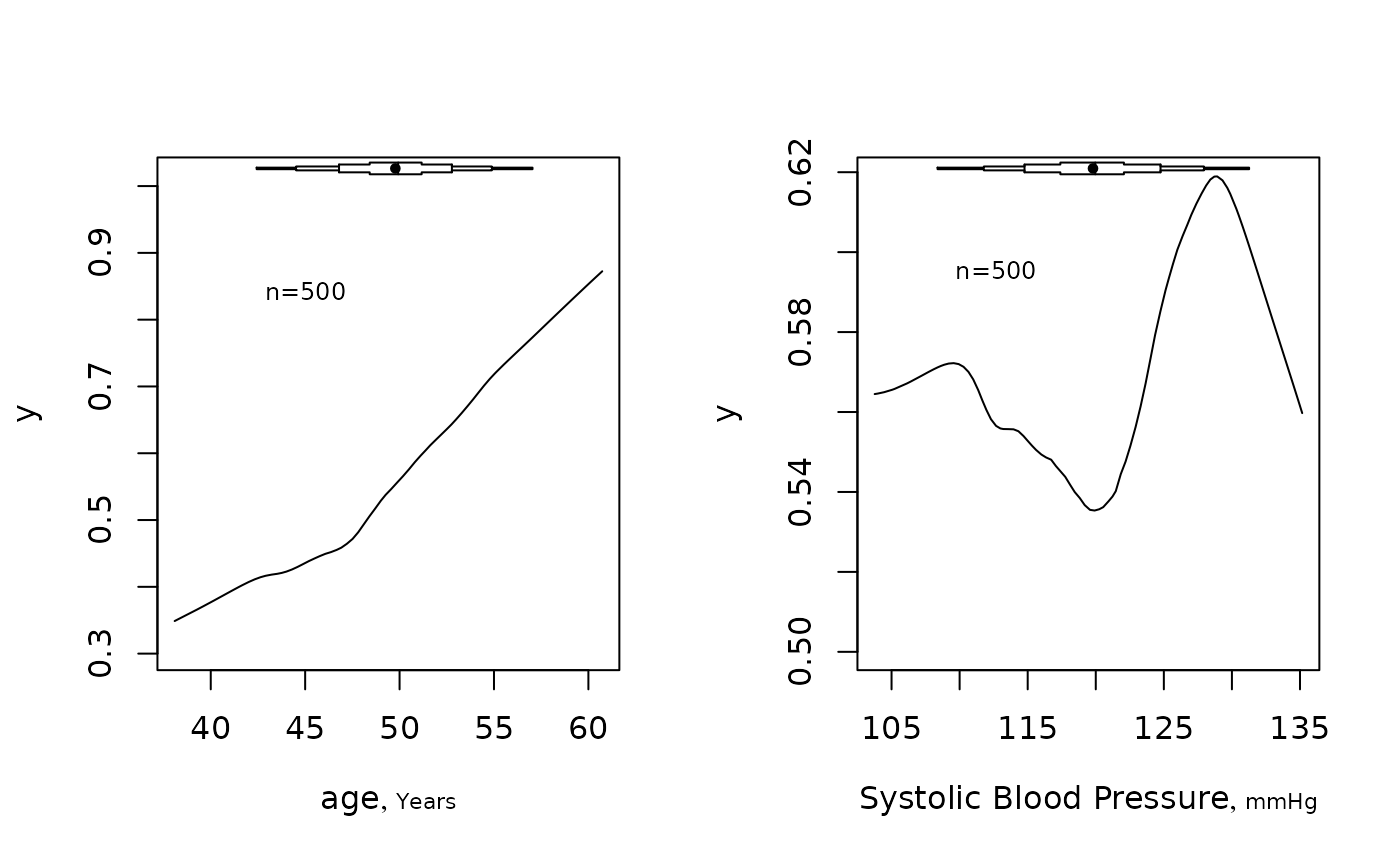
summaryRc(y ~ age + bp)
 # For x limits use 1st and 99th percentiles to frame extended box plots
summaryRc(y ~ age + bp, bpplot='top', datadensity=FALSE, trim=.01)
# For x limits use 1st and 99th percentiles to frame extended box plots
summaryRc(y ~ age + bp, bpplot='top', datadensity=FALSE, trim=.01)
 summaryRc(y ~ age + bp + stratify(sex),
label.curves=list(keys='lines'), nloc=list(x=.1, y=.05))
summaryRc(y ~ age + bp + stratify(sex),
label.curves=list(keys='lines'), nloc=list(x=.1, y=.05))
 y2 <- rbinom(500, 1, plogis(L + .5))
Y <- cbind(y, y2)
summaryRc(Y ~ age + bp + stratify(sex),
label.curves=list(keys='lines'), nloc=list(x=.1, y=.05))
y2 <- rbinom(500, 1, plogis(L + .5))
Y <- cbind(y, y2)
summaryRc(Y ~ age + bp + stratify(sex),
label.curves=list(keys='lines'), nloc=list(x=.1, y=.05))
