Accelerated Life Testing of Motorettes
motors.RdThe motors data frame has 40 rows and 3 columns. It describes an
accelerated life test at each of four temperatures of 10 motorettes,
and has rather discrete times.
motorsFormat
This data frame contains the following columns:
tempthe temperature (degrees C) of the test.
timethe time in hours to failure or censoring at 8064 hours (= 336 days).
censan indicator variable for death.
Source
Kalbfleisch, J. D. and Prentice, R. L. (1980) The Statistical Analysis of Failure Time Data. New York: Wiley.
taken from
Nelson, W. D. and Hahn, G. J. (1972) Linear regression of a regression relationship from censored data. Part 1 – simple methods and their application. Technometrics, 14, 247–276.
References
Venables, W. N. and Ripley, B. D. (2002) Modern Applied Statistics with S. Fourth edition. Springer.
Examples
library(survival)
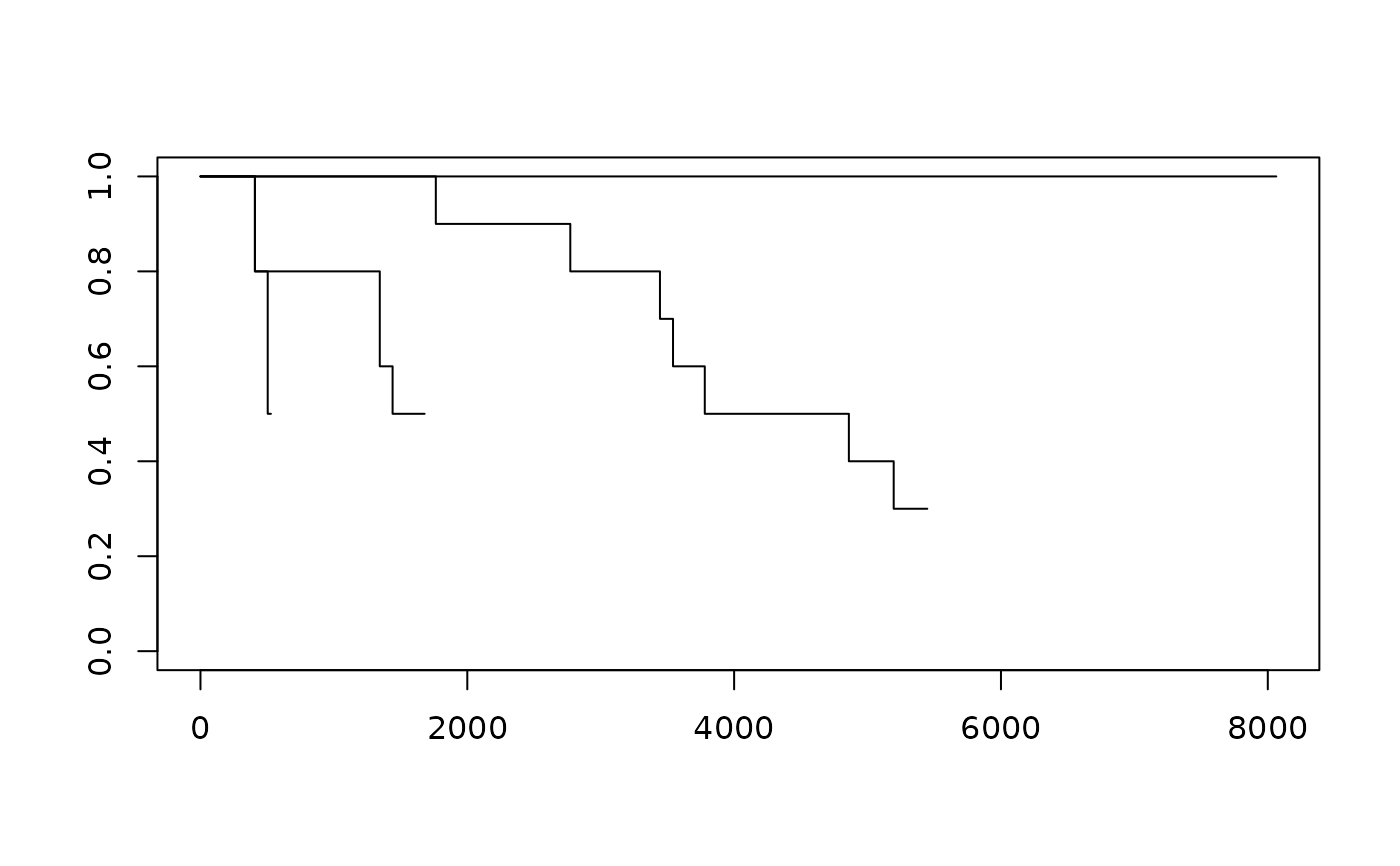
plot(survfit(Surv(time, cens) ~ factor(temp), motors), conf.int = FALSE)
 # fit Weibull model
motor.wei <- survreg(Surv(time, cens) ~ temp, motors)
## IGNORE_RDIFF_BEGIN
summary(motor.wei)
#>
#> Call:
#> survreg(formula = Surv(time, cens) ~ temp, data = motors)
#> Value Std. Error z p
#> (Intercept) 16.31852 0.62296 26.2 < 2e-16
#> temp -0.04531 0.00319 -14.2 < 2e-16
#> Log(scale) -1.09564 0.21480 -5.1 3.4e-07
#>
#> Scale= 0.334
#>
#> Weibull distribution
#> Loglik(model)= -147.4 Loglik(intercept only)= -169.5
#> Chisq= 44.32 on 1 degrees of freedom, p= 2.8e-11
#> Number of Newton-Raphson Iterations: 7
#> n= 40
#>
## IGNORE_RDIFF_END
# and predict at 130C
unlist(predict(motor.wei, data.frame(temp=130), se.fit = TRUE))
#> fit.1 se.fit.1
#> 33813.06 7506.36
motor.cox <- coxph(Surv(time, cens) ~ temp, motors)
summary(motor.cox)
#> Call:
#> coxph(formula = Surv(time, cens) ~ temp, data = motors)
#>
#> n= 40, number of events= 17
#>
#> coef exp(coef) se(coef) z Pr(>|z|)
#> temp 0.09185 1.09620 0.02736 3.358 0.000786 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> exp(coef) exp(-coef) lower .95 upper .95
#> temp 1.096 0.9122 1.039 1.157
#>
#> Concordance= 0.84 (se = 0.035 )
#> Likelihood ratio test= 25.56 on 1 df, p=4e-07
#> Wald test = 11.27 on 1 df, p=8e-04
#> Score (logrank) test = 22.73 on 1 df, p=2e-06
#>
# predict at temperature 200
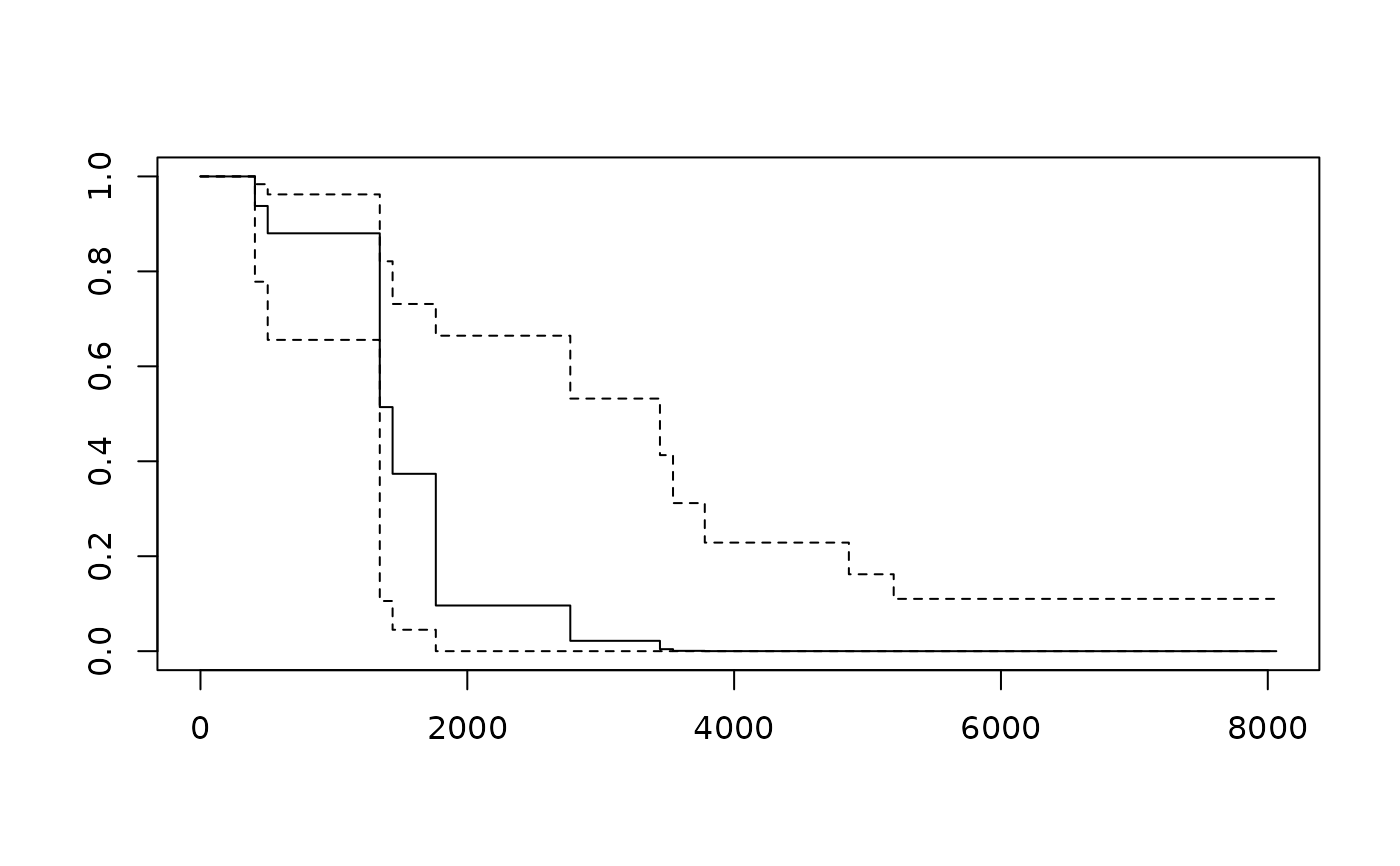
plot(survfit(motor.cox, newdata = data.frame(temp=200),
conf.type = "log-log"))
# fit Weibull model
motor.wei <- survreg(Surv(time, cens) ~ temp, motors)
## IGNORE_RDIFF_BEGIN
summary(motor.wei)
#>
#> Call:
#> survreg(formula = Surv(time, cens) ~ temp, data = motors)
#> Value Std. Error z p
#> (Intercept) 16.31852 0.62296 26.2 < 2e-16
#> temp -0.04531 0.00319 -14.2 < 2e-16
#> Log(scale) -1.09564 0.21480 -5.1 3.4e-07
#>
#> Scale= 0.334
#>
#> Weibull distribution
#> Loglik(model)= -147.4 Loglik(intercept only)= -169.5
#> Chisq= 44.32 on 1 degrees of freedom, p= 2.8e-11
#> Number of Newton-Raphson Iterations: 7
#> n= 40
#>
## IGNORE_RDIFF_END
# and predict at 130C
unlist(predict(motor.wei, data.frame(temp=130), se.fit = TRUE))
#> fit.1 se.fit.1
#> 33813.06 7506.36
motor.cox <- coxph(Surv(time, cens) ~ temp, motors)
summary(motor.cox)
#> Call:
#> coxph(formula = Surv(time, cens) ~ temp, data = motors)
#>
#> n= 40, number of events= 17
#>
#> coef exp(coef) se(coef) z Pr(>|z|)
#> temp 0.09185 1.09620 0.02736 3.358 0.000786 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> exp(coef) exp(-coef) lower .95 upper .95
#> temp 1.096 0.9122 1.039 1.157
#>
#> Concordance= 0.84 (se = 0.035 )
#> Likelihood ratio test= 25.56 on 1 df, p=4e-07
#> Wald test = 11.27 on 1 df, p=8e-04
#> Score (logrank) test = 22.73 on 1 df, p=2e-06
#>
# predict at temperature 200
plot(survfit(motor.cox, newdata = data.frame(temp=200),
conf.type = "log-log"))
 summary( survfit(motor.cox, newdata = data.frame(temp=130)) )
#> Call: survfit(formula = motor.cox, newdata = data.frame(temp = 130))
#>
#> time n.risk n.event survival std.err lower 95% CI upper 95% CI
#> 408 40 4 1.000 0.000254 0.999 1
#> 504 36 3 1.000 0.000498 0.999 1
#> 1344 28 2 0.999 0.001910 0.995 1
#> 1440 26 1 0.998 0.002697 0.993 1
#> 1764 20 1 0.996 0.005325 0.986 1
#> 2772 19 1 0.994 0.007920 0.978 1
#> 3444 18 1 0.991 0.010673 0.971 1
#> 3542 17 1 0.988 0.013667 0.962 1
#> 3780 16 1 0.985 0.016976 0.952 1
#> 4860 15 1 0.981 0.020692 0.941 1
#> 5196 14 1 0.977 0.024941 0.929 1
summary( survfit(motor.cox, newdata = data.frame(temp=130)) )
#> Call: survfit(formula = motor.cox, newdata = data.frame(temp = 130))
#>
#> time n.risk n.event survival std.err lower 95% CI upper 95% CI
#> 408 40 4 1.000 0.000254 0.999 1
#> 504 36 3 1.000 0.000498 0.999 1
#> 1344 28 2 0.999 0.001910 0.995 1
#> 1440 26 1 0.998 0.002697 0.993 1
#> 1764 20 1 0.996 0.005325 0.986 1
#> 2772 19 1 0.994 0.007920 0.978 1
#> 3444 18 1 0.991 0.010673 0.971 1
#> 3542 17 1 0.988 0.013667 0.962 1
#> 3780 16 1 0.985 0.016976 0.952 1
#> 4860 15 1 0.981 0.020692 0.941 1
#> 5196 14 1 0.977 0.024941 0.929 1