Plot model coefficients
coefplot.RdProduce dot-and-whisker plot of the model(-averaged) coefficients, with confidence intervals
coefplot(
x, lci, uci,
labels = NULL, width = 0.15,
shift = 0, horizontal = TRUE,
main = NULL, xlab = NULL, ylab = NULL,
xlim = NULL, ylim = NULL,
labAsExpr = TRUE, mar.adj = TRUE, lab.line = 0.5,
lty = par("lty"), lwd = par("lwd"), pch = 21,
col = par("col"), bg = par("bg"),
dotcex = par("cex"), dotcol = col,
staplelty = lty, staplelwd = lwd, staplecol = col,
zerolty = "dotted", zerolwd = lwd, zerocol = "gray",
las = 2, ann = TRUE, axes = TRUE, add = FALSE,
type = "p",
...
)
# S3 method for class 'averaging'
plot(
x,
full = TRUE, level = 0.95, intercept = TRUE,
parm = NULL, labels = NULL, width = 0.1,
shift = max(0.2, width * 2.1 + 0.05),
horizontal = TRUE,
xlim = NULL, ylim = NULL,
main = "Model-averaged coefficients",
xlab = NULL, ylab = NULL,
add = FALSE,
...
)Arguments
- x
either a (possibly named) vector of coefficients (for
coefplot), or an "averaging"=model.avg object.- lci, uci
vectors of lower and upper confidence intervals. Alternatively a two-column matrix with columns containing confidence intervals, in which case
uciis ignored.- labels
optional vector of coefficient names. By default, names of
xare used for labels.- width
width of the staples (= end of whisker).
- shift
the amount of perpendicular shift for the dots and whiskers. Useful when adding to an existing plot.
- horizontal
logical indicating if the plots should be horizontal; defaults to
TRUE.- main
an overall title for the plot: see
title.- xlab, ylab
x- and y-axis annotation. Can be suppressed by
ann=FALSE.- xlim, ylim
optional, the x and y limits of the plot.
- labAsExpr
logical indicating whether the coefficient names should be transformed to expressions to create prettier labels (see plotmath)
- mar.adj
logical indicating whether the (left or lower) margin should be expanded to fit the labels
- lab.line
margin line for the labels
- lty, lwd, pch, col, bg
default line type, line width, point character, foreground colour for all elements, and background colour for open symbols.
- dotcex, dotcol
dots point size expansion and colour.
- staplelty, staplelwd, staplecol
staple line type, width, and colour.
- zerolty, zerolwd, zerocol
zero-line type, line width, colour. Setting
zeroltytoNAsuppresses the line.- las
the style of labels for coefficient names. See par.
- ann
logicalindicating if axes should be annotated (byxlabandylab).- axes
a logical value indicating whether both axes should be drawn on the plot.
- add
logical, if true add to current plot.
- type
if
"n", the plot region is left empty, any other value causes the plot being drawn.- ...
additional arguments passed to
coefplotor more graphical parameters.- full
a logical value specifying whether the “full” model-averaged coefficients are plotted. If
FALSE, the “subset”-averaged coefficients are plotted, and both types ifNA. See model.avg.- level
the confidence level required.
- intercept
logical indicating if intercept should be included in the plot
- parm
a specification of which parameters are to be plotted, either a vector of numbers or a vector of names. If missing, all parameters are considered.
Value
An invisible matrix containing coordinates of points and whiskers, or,
a two-element list of such, one for each coefficient type in
plot.averaging when full is NA.
Details
Plot model(-averaged) coefficients with confidence intervals.
Examples
fm <- glm(Prop ~ dose + I(dose^2) + log(dose) + I(log(dose)^2),
data = Beetle, family = binomial, na.action = na.fail)
ma <- model.avg(dredge(fm))
#> Fixed term is "(Intercept)"
# default coefficient plot:
plot(ma, full = NA, intercept = FALSE)
 # Add colours per coefficient type
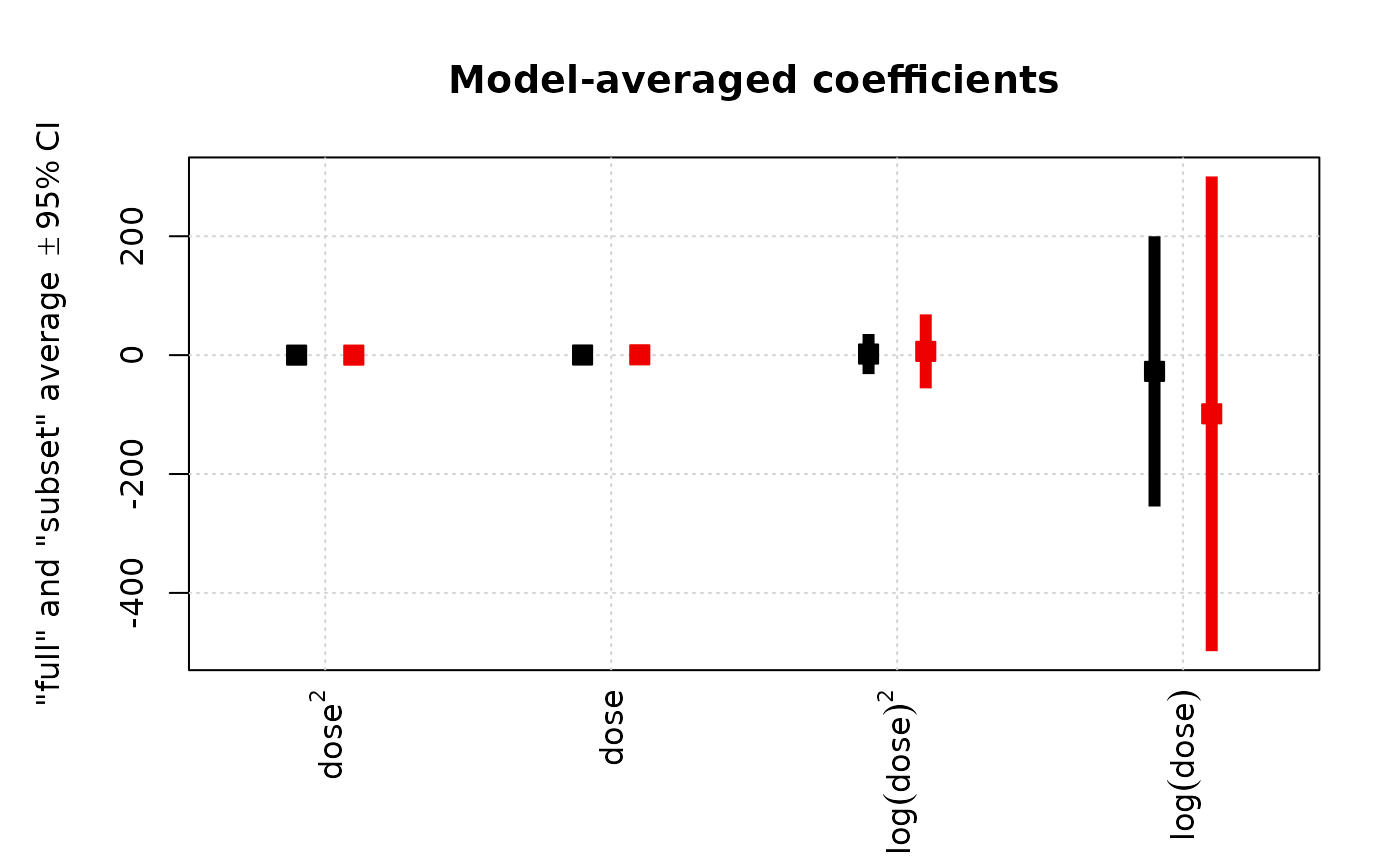
# Replicate each colour n(=number of coefficients) times
clr <- c("black", "red2")
i <- rep(1:2, each = length(coef(ma)) - 1)
plot(ma, full = NA, intercept = FALSE,
pch = 22, dotcex = 1.5,
col = clr[i], bg = clr[i],
lwd = 6, lend = 1, width = 0, horizontal = 0)
# Add colours per coefficient type
# Replicate each colour n(=number of coefficients) times
clr <- c("black", "red2")
i <- rep(1:2, each = length(coef(ma)) - 1)
plot(ma, full = NA, intercept = FALSE,
pch = 22, dotcex = 1.5,
col = clr[i], bg = clr[i],
lwd = 6, lend = 1, width = 0, horizontal = 0)
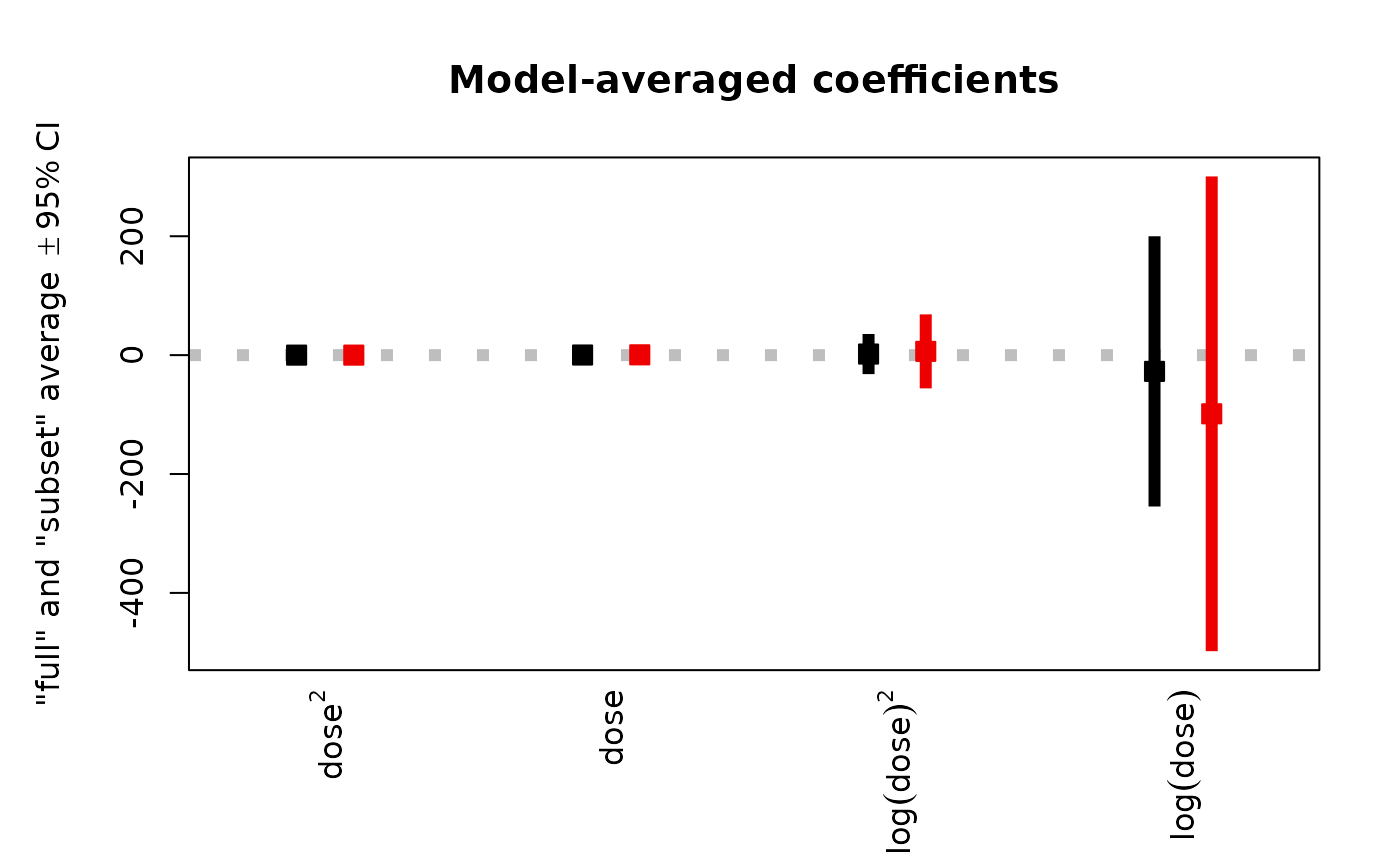 # Use `type = "n"` and `add` argument to e.g. add grid beneath the figure
plot(ma, full = NA, intercept = FALSE,
width = 0, horizontal = FALSE, zerolty = NA, type = "n")
grid()
plot(ma, full = NA, intercept = FALSE,
pch = 22, dotcex = 1.5,
col = clr[i], bg = clr[i],
lwd = 6, lend = 1, width = 0, horizontal = FALSE, add = TRUE)
# Use `type = "n"` and `add` argument to e.g. add grid beneath the figure
plot(ma, full = NA, intercept = FALSE,
width = 0, horizontal = FALSE, zerolty = NA, type = "n")
grid()
plot(ma, full = NA, intercept = FALSE,
pch = 22, dotcex = 1.5,
col = clr[i], bg = clr[i],
lwd = 6, lend = 1, width = 0, horizontal = FALSE, add = TRUE)