Visualize model selection table
plot.model.selection.RdProduces a graphical representation of model weights and terms.
# S3 method for class 'model.selection'
plot(
x,
ylab = NULL, xlab = NULL, main = "Model selection table",
labels = NULL, terms = NULL, labAsExpr = TRUE,
vlabels = rownames(x), mar.adj = TRUE,
col = NULL, col.mode = 2,
bg = "white", border = par("col"),
par.lab = NULL, par.vlab = NULL,
axes = TRUE, ann = TRUE,
...
)Arguments
- x
a
"model.selection"object.- xlab, ylab, main
labels for the x and y axes, and the main title for the plot.
- labels
optional, a character vector or an expression containing model term labels (to appear on top side of the plot). Its length must be equal to number of displayed model terms. Defaults to the model term names.
- terms
which terms to include (default
NULLmeans all terms).- labAsExpr
logical, indicating whether the term names should be interpreted (parsed) as R expressions for prettier labels. See also plotmath.
- vlabels
alternative labels for the table rows (i.e. model names)
- mar.adj
logical indicating whether the top and right margin should be enlarged if necessary to fit the labels.
- col
vector or a
matrixof colours for the non-empty grid cells. See 'Details'. Ifcolis given as a matrix, the colours are applied to rows and columns. How it is done is governed by the argumentcol.mode.- col.mode
either numeric or
"value", specifies cell colouring mode. See 'Details'.- bg
background colour for the empty cells.
- border
border colour for cells and axes.
- par.lab, par.vlab
optional lists of arguments and graphical parameters for drawing term labels (top axis) and model names (right axis), respectively. Items of
par.labare passed as arguments to mtext, and those ofpar.vlabare passed to axis.- axes, ann
logical values indicating whether the axis and annotation should appear on the plot.
- ...
further graphical parameters to be set for the plot.
Details
Colours
If col.mode = 0, the colours are recycled: if col is a matrix,
recycling takes place both per row and per column. If col.mode > 0, the
colour values in the columns are interpolated and assigned according to
the model weights. Higher values shift the colours for models with lower
model weights more forward. See also colorRamp. If col.mode < 0 or
"value" (partially matched, case-insensitive) and col has two or more
elements, colours are used to represent coefficient values: the first
element in col is used for categorical predictors, the rest for
continuous values. The default is grey for factors and
HCL palette "Blue-Red 3" otherwise, ranging from blue
for negative values to red for positive ones.
The following arguments are useful for adjusting label size and
position in par.lab and par.vlab : cex, las (see par),
line and hadj (see mtext and axis).
See also
plot.default, par, MuMIn-package
Examples
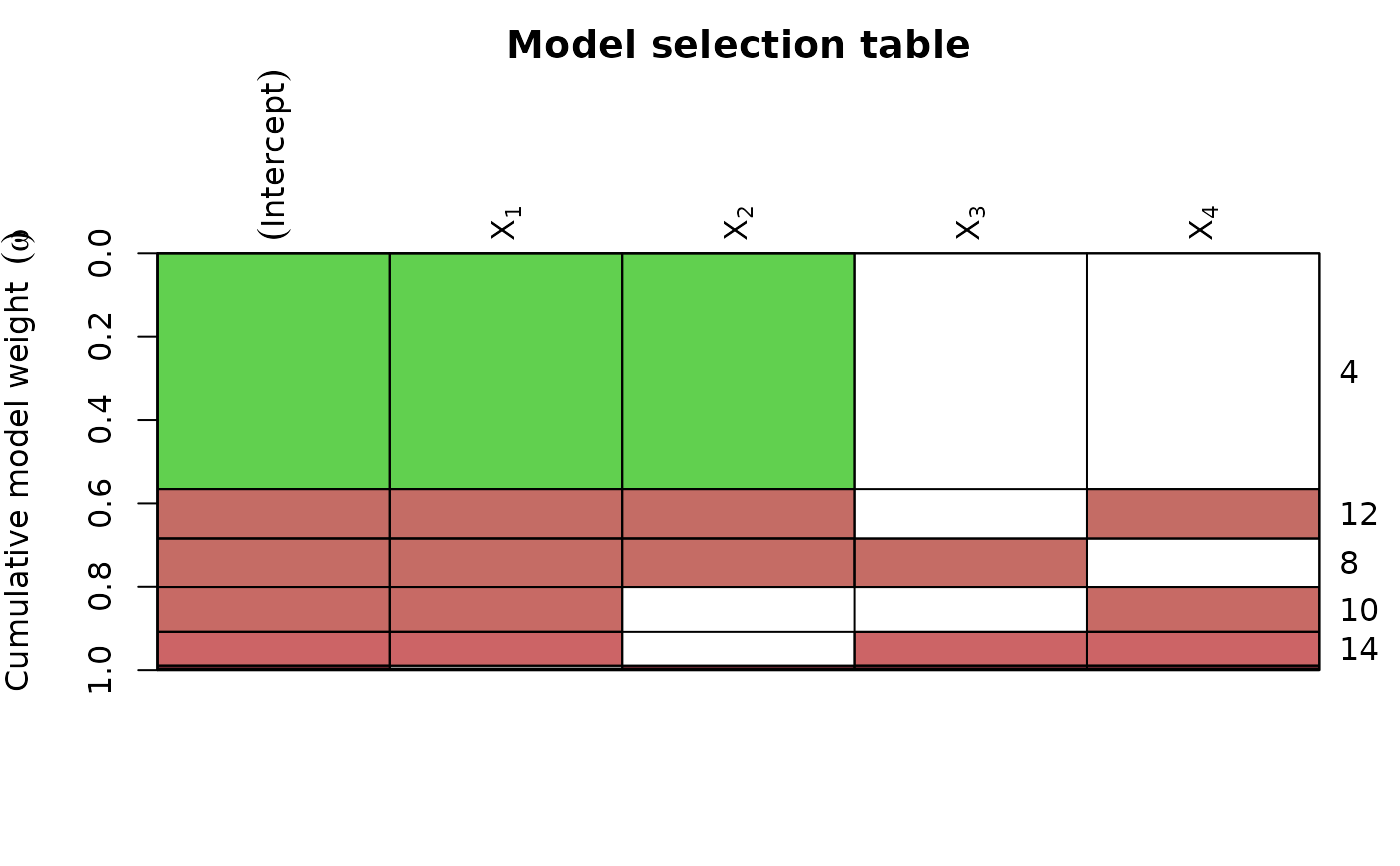
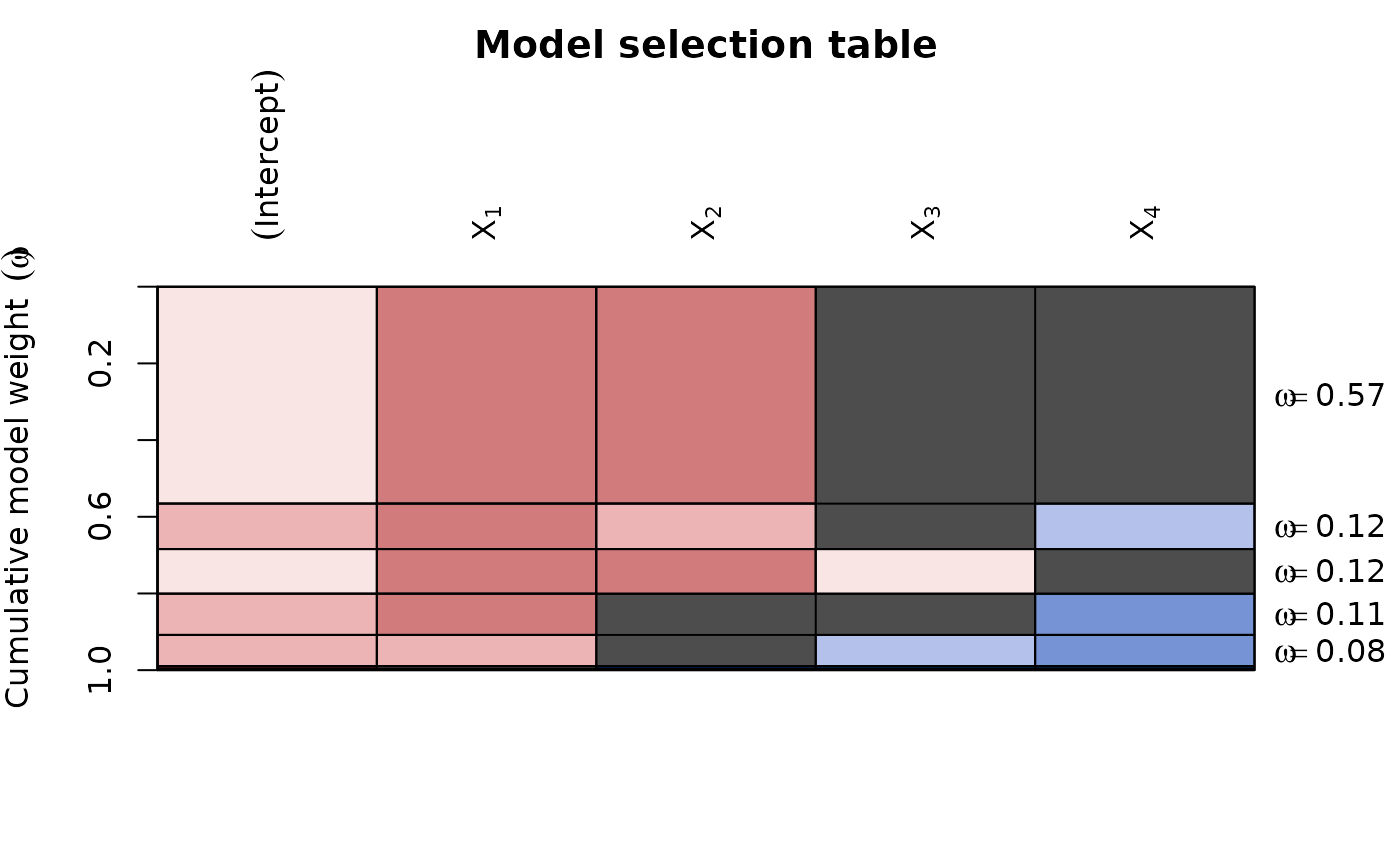
ms <- dredge(lm(formula = y ~ ., data = Cement, na.action = na.fail))
#> Fixed term is "(Intercept)"
plot(ms,
# colours by coefficient value:
col.mode = "value",
par.lab = list(las = 2, line = 1.2, cex = 1),
bg = "gray30",
# change labels for the models to Akaike weights:
vlabels = parse(text = paste("omega ==", round(Weights(ms), 2)))
)
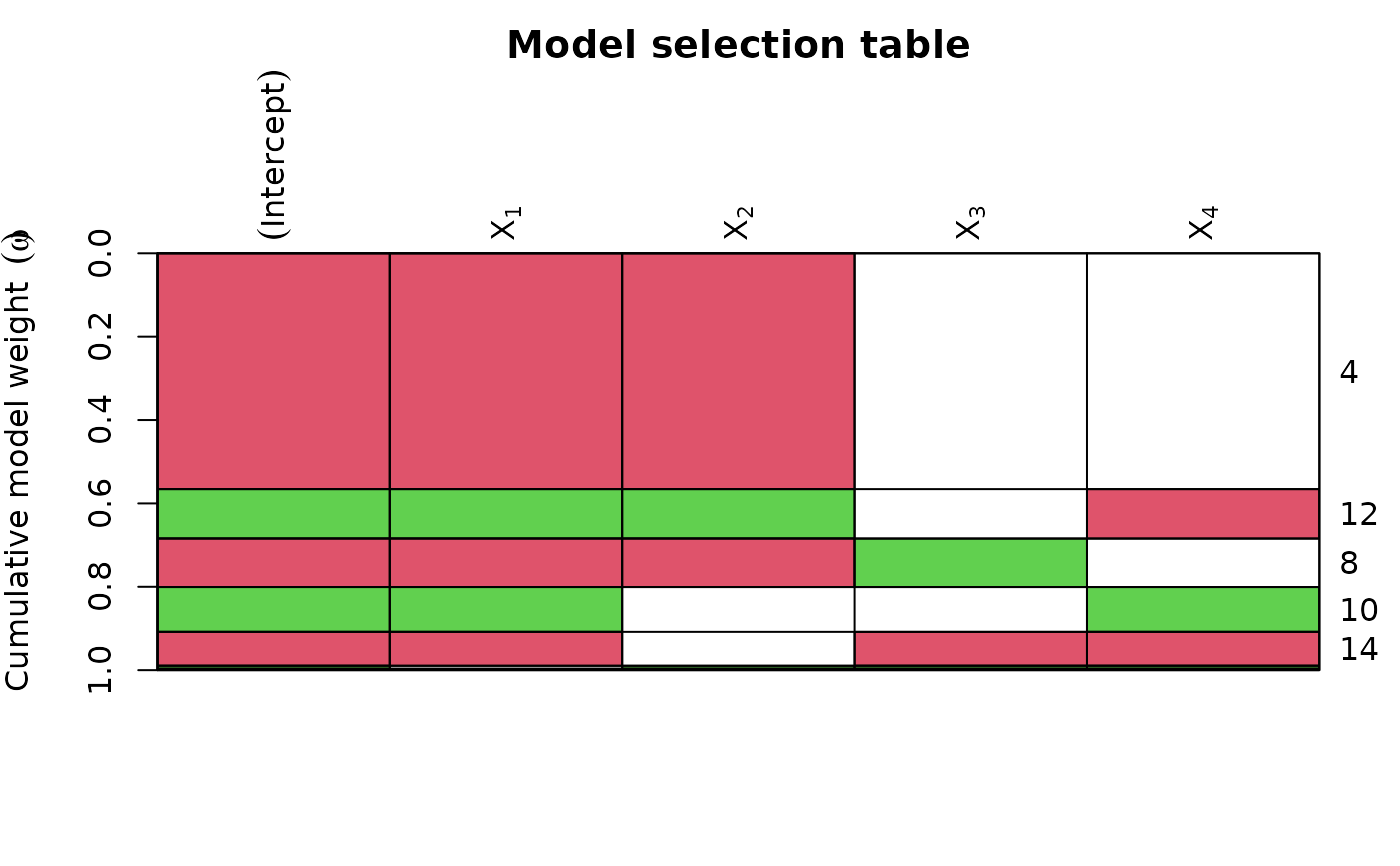
 plot(ms, col = 2:3, col.mode = 0) # colour recycled by row
plot(ms, col = 2:3, col.mode = 0) # colour recycled by row
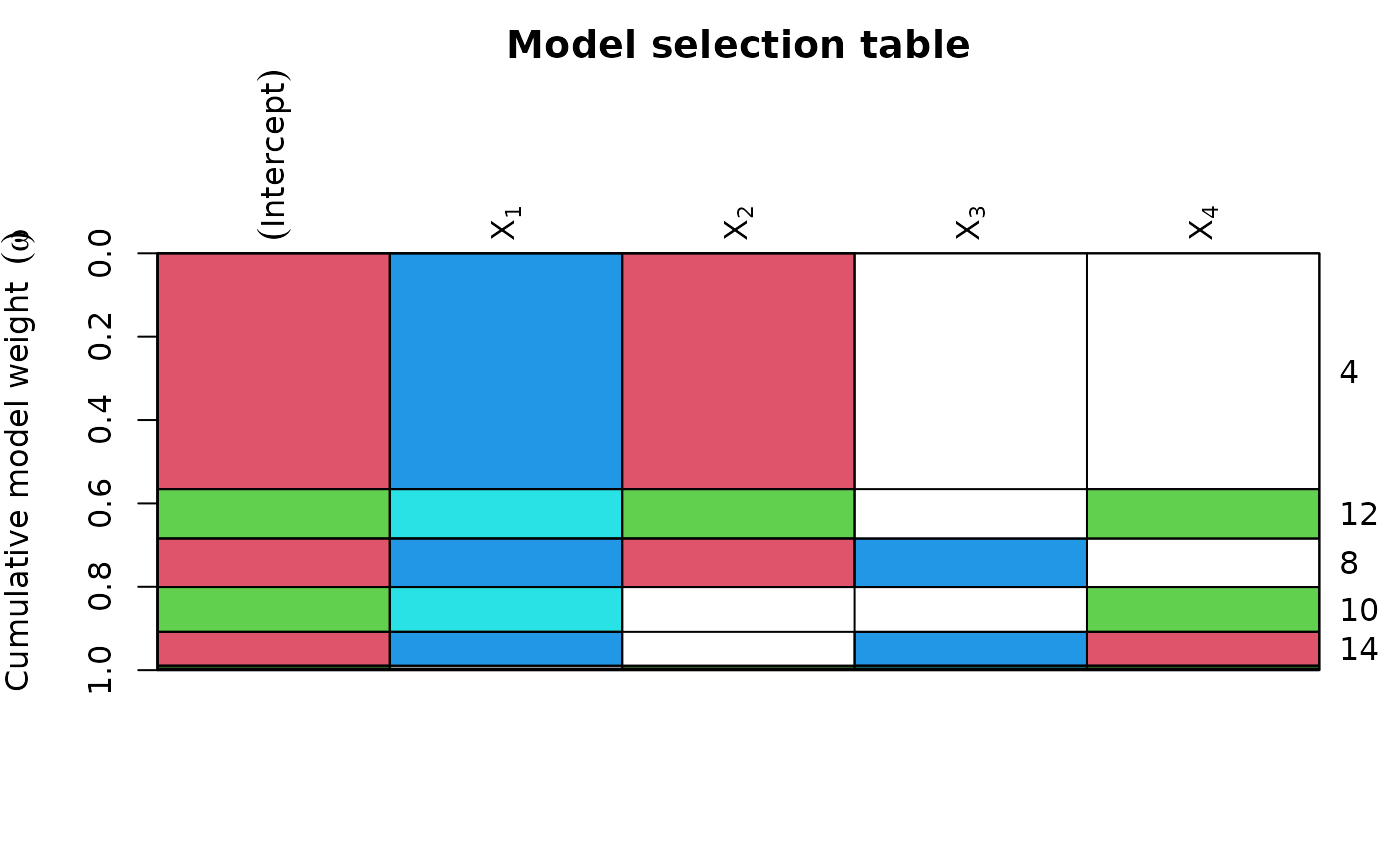
 plot(ms, col = cbind(2:3, 4:5), col.mode = 0) # colour recycled by row and column
plot(ms, col = cbind(2:3, 4:5), col.mode = 0) # colour recycled by row and column
 plot(ms, col = 2:3, col.mode = 1) # colour gradient by model weight
plot(ms, col = 2:3, col.mode = 1) # colour gradient by model weight