Restricted Regression Quantiles
rrq.RdThis function fits a restricted quantile regression model to avoid crossing of quantile curves.
rrq(formula, tau, data, subset, weights, na.action, method = "fn",
model = TRUE, contrasts = NULL, ...)
rrq.fit(x, y, tau, method = "fn", ...)
rrq.wfit(x, y, tau, weights, method = "fn", ...)Arguments
- formula
a formula object, with the response on the left of a
~operator, and the terms, separated by+operators, on the right.- x
the design matrix.
- y
the response variable.
- tau
the quantile(s) to be estimated.
- data
a data frame in which to interpret the variables named in the formula.
- subset
an optional vector specifying a subset of observations to be used in the fitting process.
- weights
an optional vector of weights to be used in the fitting process. Should be NULL or a numeric vector.
- na.action
a function which indicates what should happen when the data contain
NAs.- method
the algorithm used to compute the fit (see
rq).- model
if
TRUEthen the model frame is returned. This is essential if one wants to call summary subsequently.- contrasts
a list giving contrasts for some or all of the factors default = NULL appearing in the model formula. The elements of the list should have the same name as the variable and should be either a contrast matrix (specifically, any full-rank matrix with as many rows as there are levels in the factor), or else a function to compute such a matrix given the number of levels.
- ...
optional arguments passed to
rq.fitorrq.wfit.
References
He X. Quantile curves without crossing. The American Statistician 1997;51(2):186-192.
Koenker R. quantreg: Quantile Regression. 2016. R package version 5.29.
Examples
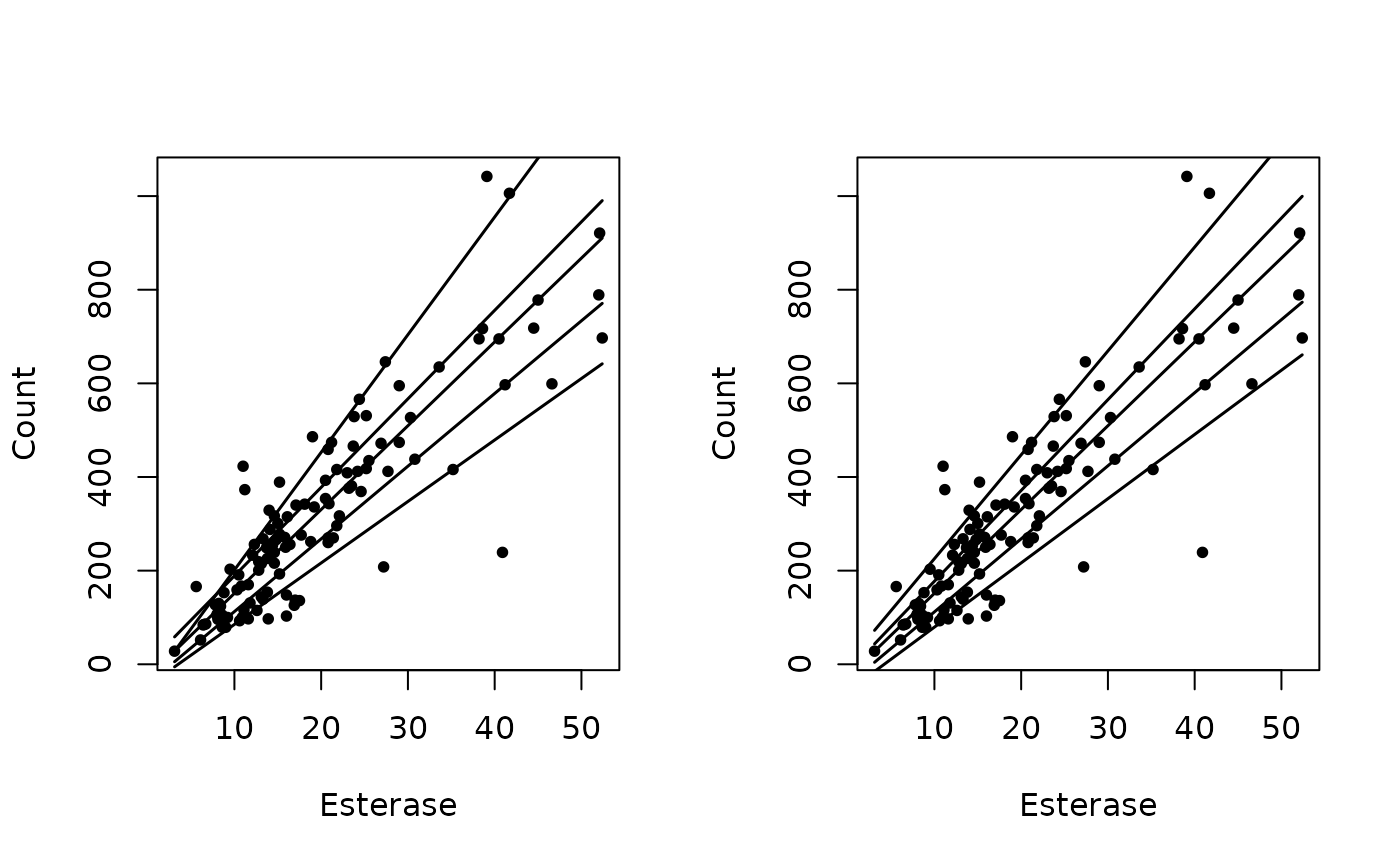
data(esterase)
# Fit standard quantile regression
fit <- quantreg::rq(Count ~ Esterase, data = esterase, tau = c(.1,.25,.5,.75,.9))
yhat <- fit$fitted.values
# Fit restricted quantile regression
fitr <- rrq(Count ~ Esterase, data = esterase, tau = c(.1,.25,.5,.75,.9))
yhat2 <- predict(fitr)
# Plot results
par(mfrow = c(1, 2))
# Plot regression quantiles
with(esterase, plot(Count ~ Esterase, pch = 16, cex = .8))
apply(yhat, 2, function(y,x) lines(x,y,lwd = 1.5), x = esterase$Esterase)
#> NULL
# Plot restricted regression quantiles
with(esterase, plot(Count ~ Esterase, pch = 16, cex = .8))
apply(yhat2, 2, function(y,x) lines(x,y,lwd = 1.5), x = esterase$Esterase)
 #> NULL
#> NULL