Quantile Regression Transformation Models
tsrq.RdThese functions are used to fit quantile regression transformation models.
tsrq(formula, data = sys.frame(sys.parent()), tsf = "mcjI", symm = TRUE,
dbounded = FALSE, lambda = NULL, conditional = FALSE, tau = 0.5,
subset, weights, na.action, contrasts = NULL, method = "fn")
tsrq2(formula, data = sys.frame(sys.parent()), dbounded = FALSE, lambda = NULL,
delta = NULL, conditional = FALSE, tau = 0.5, subset, weights, na.action,
contrasts = NULL, method = "fn")
rcrq(formula, data = sys.frame(sys.parent()), tsf = "mcjI", symm = TRUE,
dbounded = FALSE, lambda = NULL, tau = 0.5, subset, weights, na.action,
contrasts = NULL, method = "fn")
nlrq1(formula, data = sys.frame(sys.parent()), tsf = "mcjI", symm = TRUE,
dbounded = FALSE, start = NULL, tau = 0.5,
subset, weights, na.action, contrasts = NULL, control = list())
nlrq2(formula, data = sys.frame(sys.parent()), dbounded = FALSE,
start = NULL, tau = 0.5, subset, weights, na.action, contrasts = NULL)Arguments
- formula
an object of class "
formula" (or one that can be coerced to that class): a symbolic description of the model to be fitted. The details of model specification are given under `Details'.- data
an optional data frame, list or environment (or object coercible by as.data.frame to a data frame) containing the variables in the model. By default the variables are taken from the environment from which the call is made.
- tsf
transformation to be used. Possible options are
mcjIfor Proposal I transformation models (default),bcfor Box-Cox andaofor Aranda-Ordaz transformation models.- symm
logical flag. If
TRUE(default) a symmetric transformation is used.- dbounded
logical flag. If
TRUEthe response is assumed to be doubly bounded on [a,b]. IfFALSE(default) the response is assumed to be singly bounded (ie, strictly positive).- lambda, delta
values of transformation parameters for grid search.
- conditional
logical flag. If
TRUE, the transformation parameter is assumed to be known and this must be provided via the argumentslambda,deltain vectors of the same length astau.- start
vector of length
1 + p(nlrq1) or2 + p(nlrq2) of initial values for the parameters to be optimized over. The first one (nlrq1) or two (nlrq2) values for the transformation parameterlambda, orlambdaanddelta, while the lastpvalues are for the regression coefficients. These initial values are passed tonl.fit.rqtor tooptim.- control
list of control parameters of the fitting process (nlrq1). See
nlControl.- tau
the quantile(s) to be estimated. See
rq.- subset
an optional vector specifying a subset of observations to be used in the fitting process.
- weights
an optional vector of weights to be used in the fitting process. Should be NULL or a numeric vector.
- na.action
a function which indicates what should happen when the data contain
NAs.- contrasts
an optional list. See the
contrasts.argofmodel.matrix.default.- method
fitting algorithm for
rq(default is Frisch-Newton interior point method "fn").
Details
These functions implement quantile regression transformation models as discussed by Geraci and Jones (see references). The general model is assumed to be \(Q_{h(Y)|X}(\tau) = \eta = Xb\), where \(Q\) denotes the conditional quantile function, \(Y\) is the response variable, \(X\) a design matrix, and \(h\) is a monotone one- or two-parameter transformation. A typical model specified in formula has the form response ~ terms where response is the (numeric) response vector and terms is a series of terms which specifies a linear predictor for the quantile of the transformed response. The response, which is singly or doubly bounded, i.e. response > 0 or 0 <= response <= 1 respectively, undergoes the transformation specified in tsf. If the response is bounded in the generic \([a,b]\) interval, the latter is automatically mapped to \([0,1]\) and no further action is required. If, however, the response is singly bounded and contains negative values, it is left to the user to offset the response or the code will produce an error.
The functions tsrq and tsrq2 use a two-stage (TS) estimator (Fitzenberger et al, 2010) for, respectively, one- and two-parameter transformations. The function rcrq (one-parameter tranformations) is based on the residual cusum process estimator proposed by Mu and He (2007). The functions nlrq1 (one-parameter tranformations) and nlrq2 (two-parameter tranformations) are based on, respectively, gradient search and Nelder-Mead optimization.
Value
tsrq, tsrq2, rcrq, nlrq2 return an object of class rqt. This is a list that contains as typical components:
- ...
the first
nt = length(tau)elements of the list store the results from fitting linear quantile models on the tranformed scale of the response.- call
the matched call.
- method
the fitting algorithm for
rqoroptim.- y
the response – untransformed scale.
- theta
if
dbounded = TRUE, the response mapped to the unit interval.- x
the model matrix.
- weights
the weights used in the fitting process (a vector of 1's if
weightsis missing orNULL).- tau
the order of the estimated quantile(s).
- lambda
the estimated parameter lambda.
- eta
the estimated parameters lambda and delta in the two-parameter Proposal II tranformation.
- lambda.grid
grid of lambda values used for estimation.
- delta.grid
grid of delta values used for estimation.
- tsf
tranformation used (see also
attributes(tsf)).- objective
values of the objective function minimised over the tranformation parameter(s). This is an array of dimension
c(nl,nt)orc(nl,nd,nt), wherenl = length(lambda.grid),nd = length(delta.grid)andnt = length(tau).- optimum
value of the objective function at solution.
- coefficients
quantile regression coefficients – transformed scale.
- fitted.values
fitted values.
- rejected
proportion of inadmissible observations (Fitzenberger et al, 2010).
- terms
the
termsused.- term.labels
names of coefficients.
- rdf
residual degrees of freedom.
References
Aranda-Ordaz FJ. On two families of transformations to additivity for binary response data. Biometrika 1981;68(2):357-363.
Box GEP, Cox DR. An analysis of transformations. Journal of the Royal Statistical Society Series B-Statistical Methodology 1964;26(2):211-252.
Dehbi H-M, Cortina-Borja M, and Geraci M. Aranda-Ordaz quantile regression for student performance assessment. Journal of Applied Statistics. 2016;43(1):58-71.
Fitzenberger B, Wilke R, Zhang X. Implementing Box-Cox quantile regression. Econometric Reviews 2010;29(2):158-181.
Geraci M and Jones MC. Improved transformation-based quantile regression. Canadian Journal of Statistics 2015;43(1):118-132.
Jones MC. Connecting distributions with power tails on the real line, the half line and the interval. International Statistical Review 2007;75(1):58-69.
Koenker R. quantreg: Quantile Regression. 2016. R package version 5.29.
Mu YM, He XM. Power transformation toward a linear regression quantile. Journal of the American Statistical Association 2007;102(477):269-279.
See also
Examples
###########################################################
## Example 1 - singly bounded (from Geraci and Jones, 2014)
if (FALSE) { # \dontrun{
data(trees)
require(MASS)
dx <- 0.01
lambda0 <- boxcox(Volume ~ log(Height), data = trees,
lambda = seq(-0.9, 0.5, by = dx))
lambda0 <- lambda0$x[which.max(lambda0$y)]
trees$z <- bc(trees$Volume,lambda0)
trees$y <- trees$Volume
trees$x <- log(trees$Height)
trees$x <- trees$x - mean(log(trees$Height))
fit.lm <- lm(z ~ x, data = trees)
newd <- data.frame(x = log(seq(min(trees$Height),
max(trees$Height), by = 0.1)))
newd$x <- newd$x - mean(log(trees$Height))
ylm <- invbc(predict(fit.lm, newdata = newd), lambda0)
lambdas <- list(bc = seq(-10, 10, by=dx),
mcjIs = seq(0,10,by = dx), mcjIa = seq(0,20,by = dx))
taus <- 1:3/4
fit0 <- tsrq(y ~ x, data = trees, tsf = "bc", symm = FALSE,
lambda = lambdas$bc, tau = taus)
fit1 <- tsrq(y ~ x, data = trees, tsf = "mcjI", symm = TRUE,
dbounded = FALSE, lambda = lambdas$mcjIs, tau = taus)
fit2 <- tsrq(y ~ x, data = trees, tsf = "mcjI", symm = FALSE,
dbounded = FALSE, lambda = lambdas$mcjIa, tau = taus)
par(mfrow = c(1,3), mar = c(7.1, 7.1, 5.1, 2.1), mgp = c(5, 2, 0))
cx.lab <- 2.5
cx.ax <- 2
lw <- 2
cx <- 2
xb <- "log(Height)"
yb <- "Volume"
xl <- range(trees$x)
yl <- c(5,80)
yhat <- predict(fit0, newdata = newd)
plot(y ~ x, data = trees, xlim = xl, ylim = yl, main = "Box-Cox",
cex.lab = cx.lab, cex.axis = cx.ax, cex.main = cx.lab,
cex = cx, xlab = xb, ylab = yb)
lines(newd$x, yhat[,1], lwd = lw)
lines(newd$x, yhat[,2], lwd = lw)
lines(newd$x, yhat[,3], lwd = lw)
lines(newd$x, ylm, lwd = lw, lty = 2)
yhat <- predict(fit1, newdata = newd)
plot(y ~ x, data = trees, xlim = xl, ylim = yl, main = "Proposal I (symmetric)",
cex.lab = cx.lab, cex.axis = cx.ax, cex.main = cx.lab,
cex = cx, xlab = xb, ylab = yb)
lines(newd$x, yhat[,1], lwd = lw)
lines(newd$x, yhat[,2], lwd = lw)
lines(newd$x, yhat[,3], lwd = lw)
lines(newd$x, ylm, lwd = lw, lty = 2)
yhat <- predict(fit2, newdata = newd)
plot(y ~ x, data = trees, xlim = xl, ylim = yl, main = "Proposal I (asymmetric)",
cex.lab = cx.lab, cex.axis = cx.ax, cex.main = cx.lab,
cex = cx, xlab = xb, ylab = yb)
lines(newd$x, yhat[,1], lwd = lw)
lines(newd$x, yhat[,2], lwd = lw)
lines(newd$x, yhat[,3], lwd = lw)
lines(newd$x, ylm, lwd = lw, lty = 2)
} # }
###########################################################
## Example 2 - doubly bounded
if (FALSE) { # \dontrun{
data(Chemistry)
Chemistry$gcse_gr <- cut(Chemistry$gcse, c(0,seq(4,8,by=0.5)))
with(Chemistry, plot(score ~ gcse_gr, xlab = "GCSE score",
ylab = "A-level Chemistry score"))
# The dataset has > 31000 observations and computation can be slow
set.seed(178)
chemsub <- Chemistry[sample(1:nrow(Chemistry), 2000), ]
# Fit symmetric Aranda-Ordaz quantile 0.9
tsrq(score ~ gcse, data = chemsub, tsf = "ao", symm = TRUE,
lambda = seq(0,2,by=0.01), tau = 0.9)
# Fit symmetric Proposal I quantile 0.9
tsrq(score ~ gcse, data = chemsub, tsf = "mcjI", symm = TRUE,
dbounded = TRUE, lambda = seq(0,2,by=0.01), tau = 0.9)
# Fit Proposal II quantile 0.9 (Nelder-Mead)
nlrq2(score ~ gcse, data = chemsub, dbounded = TRUE, tau = 0.9)
# Fit Proposal II quantile 0.9 (grid search)
# This is slower than nlrq2 but more stable numerically
tsrq2(score ~ gcse, data = chemsub, dbounded = TRUE,
lambda = seq(0, 2, by = 0.1), delta = seq(0, 2, by = 0.1),
tau = 0.9)
} # }
###########################################################
## Example 3 - doubly bounded
data(labor)
new <- labor
new$y <- new$pain
new$x <- (new$time-30)/30
new$x_gr <- as.factor(new$x)
par(mfrow = c(2,2))
cx.lab <- 1
cx.ax <- 2.5
cx <- 2.5
yl <- c(0,0.06)
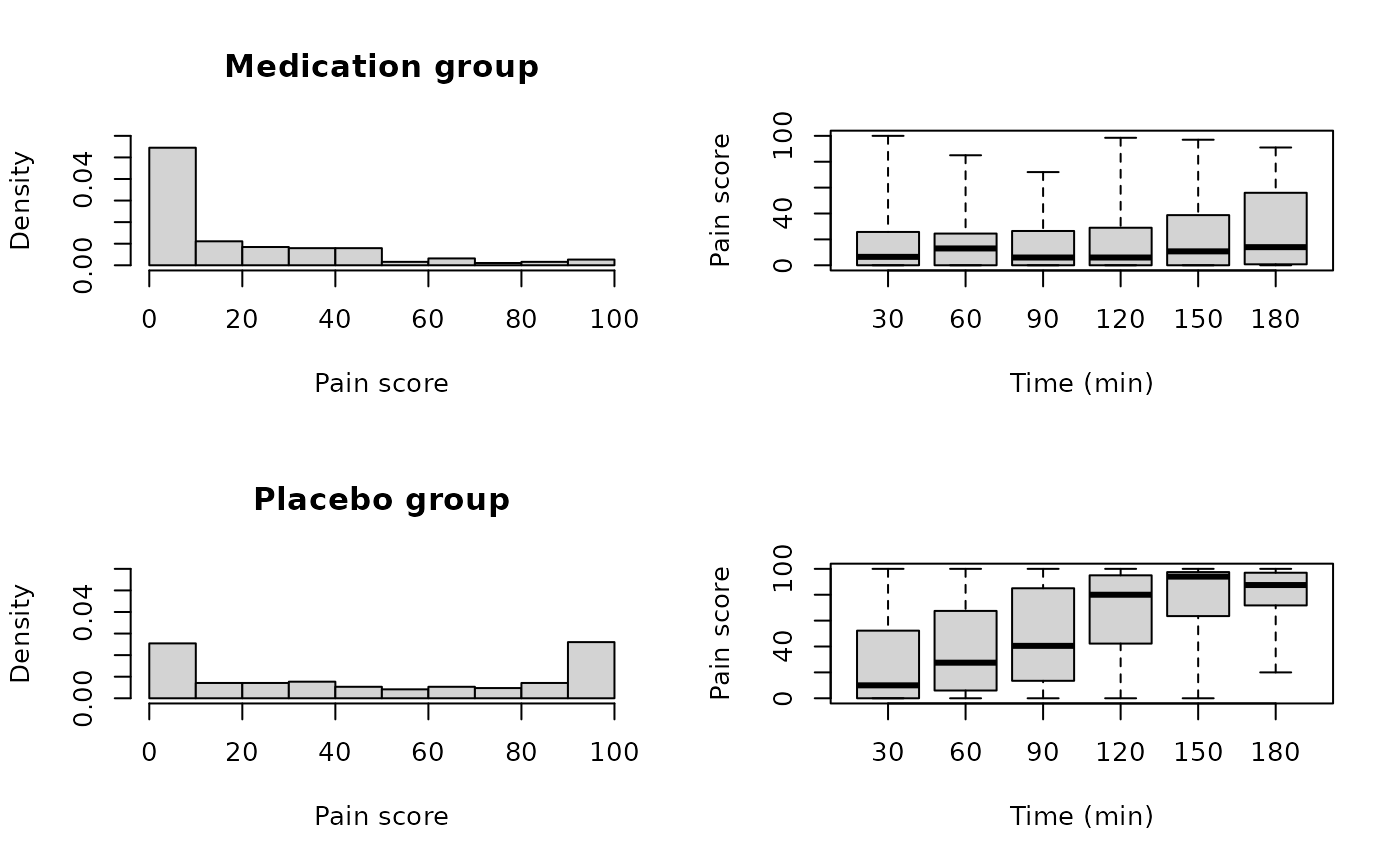
hist(new$y[new$treatment == 1], xlab = "Pain score", main = "Medication group",
freq = FALSE, ylim = yl)
plot(y ~ x_gr, new, subset = new$treatment == 1, xlab = "Time (min)",
ylab = "Pain score", axes = FALSE, range = 0)
axis(1, at = 1:6, labels = c(0:5)*30 + 30)
axis(2)
box()
hist(new$y[new$treatment == 0], xlab = "Pain score", main = "Placebo group",
freq = FALSE, ylim = yl)
plot(y ~ x_gr, new, subset = new$treatment == 0, xlab = "Time (min)",
ylab = "Pain score", axes = FALSE, range = 0)
axis(1, at = 1:6, labels = (0:5)*30 + 30)
axis(2)
box()
 #
if (FALSE) { # \dontrun{
taus <- c(1:3/4)
ls <- seq(0,3.5,by=0.1)
fit.aos <- tsrq(y ~ x*treatment, data = new, tsf = "ao", symm = TRUE,
dbounded = TRUE, tau = taus, lambda = ls)
fit.aoa <- tsrq(y ~ x*treatment, data = new, tsf = "ao", symm = FALSE,
dbounded = TRUE, tau = taus, lambda = ls)
fit.mcjs <- tsrq(y ~ x*treatment, data = new, tsf = "mcjI", symm = TRUE,
dbounded = TRUE, tau = taus, lambda = ls)
fit.mcja <- tsrq(y ~ x*treatment, data = new, tsf = "mcjI", symm = FALSE,
dbounded = TRUE, tau = taus, lambda = ls)
fit.mcj2 <- tsrq2(y ~ x*treatment, data = new, dbounded = TRUE, tau = taus,
lambda = seq(0,2,by=0.1), delta = seq(0,1.5,by=0.3))
fit.nlrq <- nlrq2(y ~ x*treatment, data = new, start = coef(fit.mcj2, all = TRUE)[,1],
dbounded = TRUE, tau = taus)
sel <- 0 # placebo (change to sel == 1 for medication group)
x <- new$x
nd <- data.frame(x = seq(min(x), max(x), length=200), treatment = sel)
xx <- nd$x+1
par(mfrow = c(2,2))
fit <- fit.aos
yhat <- predict(fit, newdata = nd)
plot(y ~ x_gr, new, subset = new$treatment == sel, xlab = "",
ylab = "Pain score", axes = FALSE, main = "Aranda-Ordaz (s)",
range = 0, col = grey(4/5))
apply(yhat, 2, function(y,x) lines(x, y, lwd = 2), x = xx)
axis(1, at = 1:6, labels = (0:5)*30 + 30)
axis(2, at = c(0, 25, 50, 75, 100))
box()
fit <- fit.aoa
yhat <- predict(fit, newdata = nd)
plot(y ~ x_gr, new, subset = new$treatment == sel, xlab = "", ylab = "",
axes = FALSE, main = "Aranda-Ordaz (a)", range = 0, col = grey(4/5))
apply(yhat, 2, function(y,x) lines(x, y, lwd = 2), x = xx)
axis(1, at = 1:6, labels = (0:5)*30 + 30)
axis(2, at = c(0, 25, 50, 75, 100))
box()
fit <- fit.mcjs
yhat <- predict(fit, newdata = nd)
plot(y ~ x_gr, new, subset = new$treatment == sel, xlab = "Time (min)",
ylab = "Pain score", axes = FALSE, main = "Proposal I (s)",
range = 0, col = grey(4/5))
apply(yhat, 2, function(y,x) lines(x, y, lwd = 2), x = xx)
axis(1, at = 1:6, labels = (0:5)*30 + 30)
axis(2, at = c(0, 25, 50, 75, 100))
box()
fit <- fit.mcj2
yhat <- predict(fit, newdata = nd)
plot(y ~ x_gr, new, subset = new$treatment == sel, xlab = "Time (min)",
ylab = "", axes = FALSE, main = "Proposal II", range = 0, col = grey(4/5))
apply(yhat, 2, function(y,x) lines(x, y, lwd = 2), x = xx)
axis(1, at = 1:6, labels = (0:5)*30 + 30)
axis(2, at = c(0, 25, 50, 75, 100))
box()
} # }
#
if (FALSE) { # \dontrun{
taus <- c(1:3/4)
ls <- seq(0,3.5,by=0.1)
fit.aos <- tsrq(y ~ x*treatment, data = new, tsf = "ao", symm = TRUE,
dbounded = TRUE, tau = taus, lambda = ls)
fit.aoa <- tsrq(y ~ x*treatment, data = new, tsf = "ao", symm = FALSE,
dbounded = TRUE, tau = taus, lambda = ls)
fit.mcjs <- tsrq(y ~ x*treatment, data = new, tsf = "mcjI", symm = TRUE,
dbounded = TRUE, tau = taus, lambda = ls)
fit.mcja <- tsrq(y ~ x*treatment, data = new, tsf = "mcjI", symm = FALSE,
dbounded = TRUE, tau = taus, lambda = ls)
fit.mcj2 <- tsrq2(y ~ x*treatment, data = new, dbounded = TRUE, tau = taus,
lambda = seq(0,2,by=0.1), delta = seq(0,1.5,by=0.3))
fit.nlrq <- nlrq2(y ~ x*treatment, data = new, start = coef(fit.mcj2, all = TRUE)[,1],
dbounded = TRUE, tau = taus)
sel <- 0 # placebo (change to sel == 1 for medication group)
x <- new$x
nd <- data.frame(x = seq(min(x), max(x), length=200), treatment = sel)
xx <- nd$x+1
par(mfrow = c(2,2))
fit <- fit.aos
yhat <- predict(fit, newdata = nd)
plot(y ~ x_gr, new, subset = new$treatment == sel, xlab = "",
ylab = "Pain score", axes = FALSE, main = "Aranda-Ordaz (s)",
range = 0, col = grey(4/5))
apply(yhat, 2, function(y,x) lines(x, y, lwd = 2), x = xx)
axis(1, at = 1:6, labels = (0:5)*30 + 30)
axis(2, at = c(0, 25, 50, 75, 100))
box()
fit <- fit.aoa
yhat <- predict(fit, newdata = nd)
plot(y ~ x_gr, new, subset = new$treatment == sel, xlab = "", ylab = "",
axes = FALSE, main = "Aranda-Ordaz (a)", range = 0, col = grey(4/5))
apply(yhat, 2, function(y,x) lines(x, y, lwd = 2), x = xx)
axis(1, at = 1:6, labels = (0:5)*30 + 30)
axis(2, at = c(0, 25, 50, 75, 100))
box()
fit <- fit.mcjs
yhat <- predict(fit, newdata = nd)
plot(y ~ x_gr, new, subset = new$treatment == sel, xlab = "Time (min)",
ylab = "Pain score", axes = FALSE, main = "Proposal I (s)",
range = 0, col = grey(4/5))
apply(yhat, 2, function(y,x) lines(x, y, lwd = 2), x = xx)
axis(1, at = 1:6, labels = (0:5)*30 + 30)
axis(2, at = c(0, 25, 50, 75, 100))
box()
fit <- fit.mcj2
yhat <- predict(fit, newdata = nd)
plot(y ~ x_gr, new, subset = new$treatment == sel, xlab = "Time (min)",
ylab = "", axes = FALSE, main = "Proposal II", range = 0, col = grey(4/5))
apply(yhat, 2, function(y,x) lines(x, y, lwd = 2), x = xx)
axis(1, at = 1:6, labels = (0:5)*30 + 30)
axis(2, at = c(0, 25, 50, 75, 100))
box()
} # }