Hat Values and Regression Deletion Diagnostics
hatvalues.RdWhen complete, a suite of functions that can be used to compute some of the regression (leave-one-out deletion) diagnostics, for the VGLM class.
hatvalues(model, ...)
hatvaluesvlm(model, type = c("diagonal", "matrix", "centralBlocks"), ...)
hatplot(model, ...)
hatplot.vlm(model, multiplier = c(2, 3), lty = "dashed",
xlab = "Observation", ylab = "Hat values", ylim = NULL, ...)
dfbetavlm(model, maxit.new = 1,
trace.new = FALSE,
smallno = 1.0e-8, ...)Arguments
- model
an R object, typically returned by
vglm.- type
Character. The default is the first choice, which is a \(nM \times nM\) matrix. If
type = "matrix"then the entire hat matrix is returned. Iftype = "centralBlocks"then \(n\) central \(M \times M\) block matrices, in matrix-band format.
- multiplier
Numeric, the multiplier. The usual rule-of-thumb is that values greater than two or three times the average leverage (at least for the linear model) should be checked.
- lty, xlab, ylab, ylim
Graphical parameters, see
paretc. The default ofylimisc(0, max(hatvalues(model)))which means that if the horizontal dashed lines cannot be seen then there are no particularly influential observations.- maxit.new, trace.new, smallno
Having
maxit.new = 1will give a one IRLS step approximation from the ordinary solution (and no warnings!). Else havingmaxit.new = 10, say, should usually mean convergence will occur for all observations when they are removed one-at-a-time. Else havingmaxit.new = 2, say, should usually mean some lack of convergence will occur when observations are removed one-at-a-time. Settingtrace.new = TRUEwill produce some running output at each IRLS iteration and for each individual row of the model matrix. The argumentsmallnomultiplies each value of the original prior weight (often unity); setting it identically to zero will result in an error, but setting a very small value effectively removes that observation.
Details
The invocation hatvalues(vglmObject) should return a
\(n \times M\) matrix of the diagonal elements of the
hat (projection) matrix of a vglm object.
To do this,
the QR decomposition of the object is retrieved or
reconstructed, and then straightforward calculations
are performed.
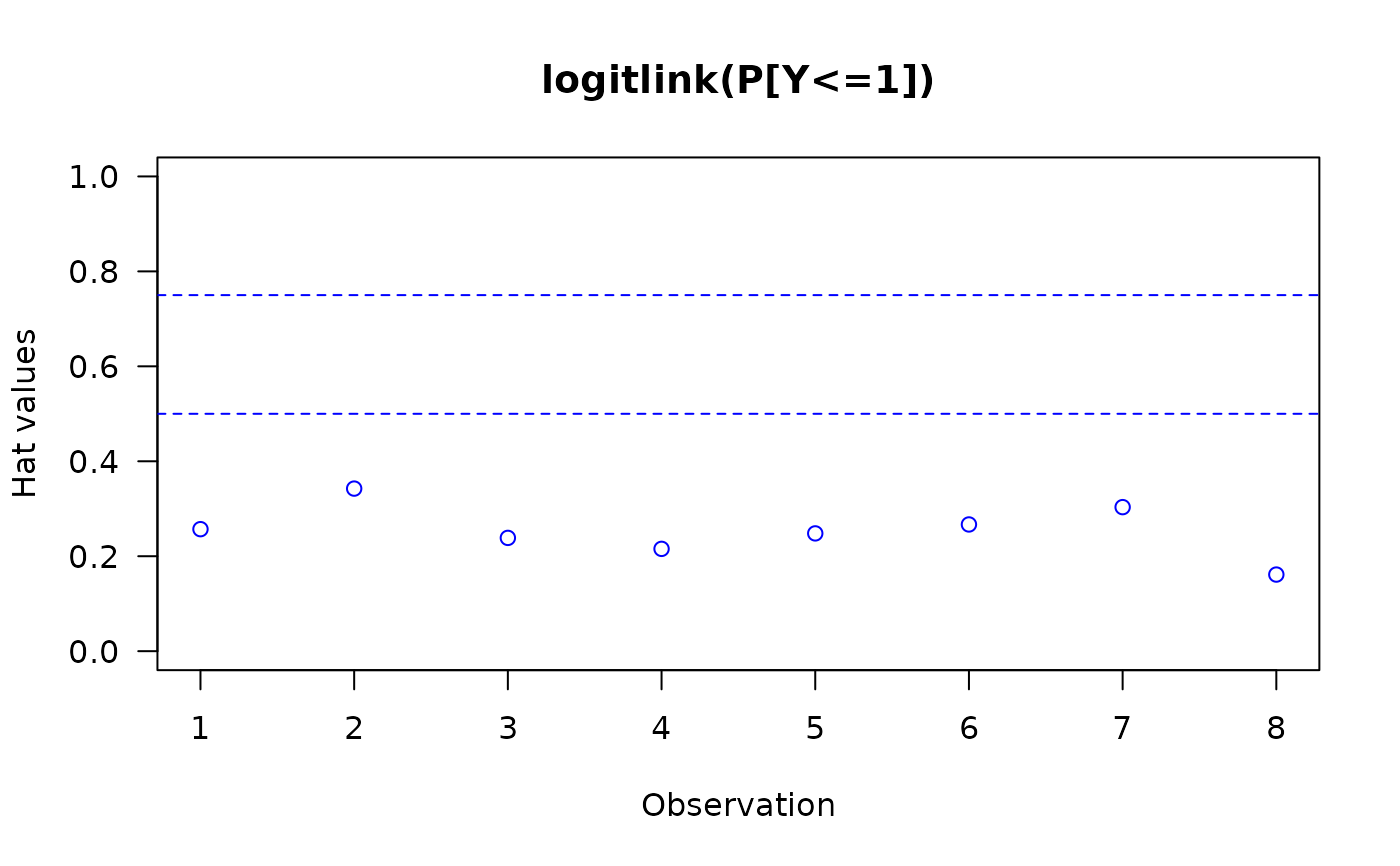
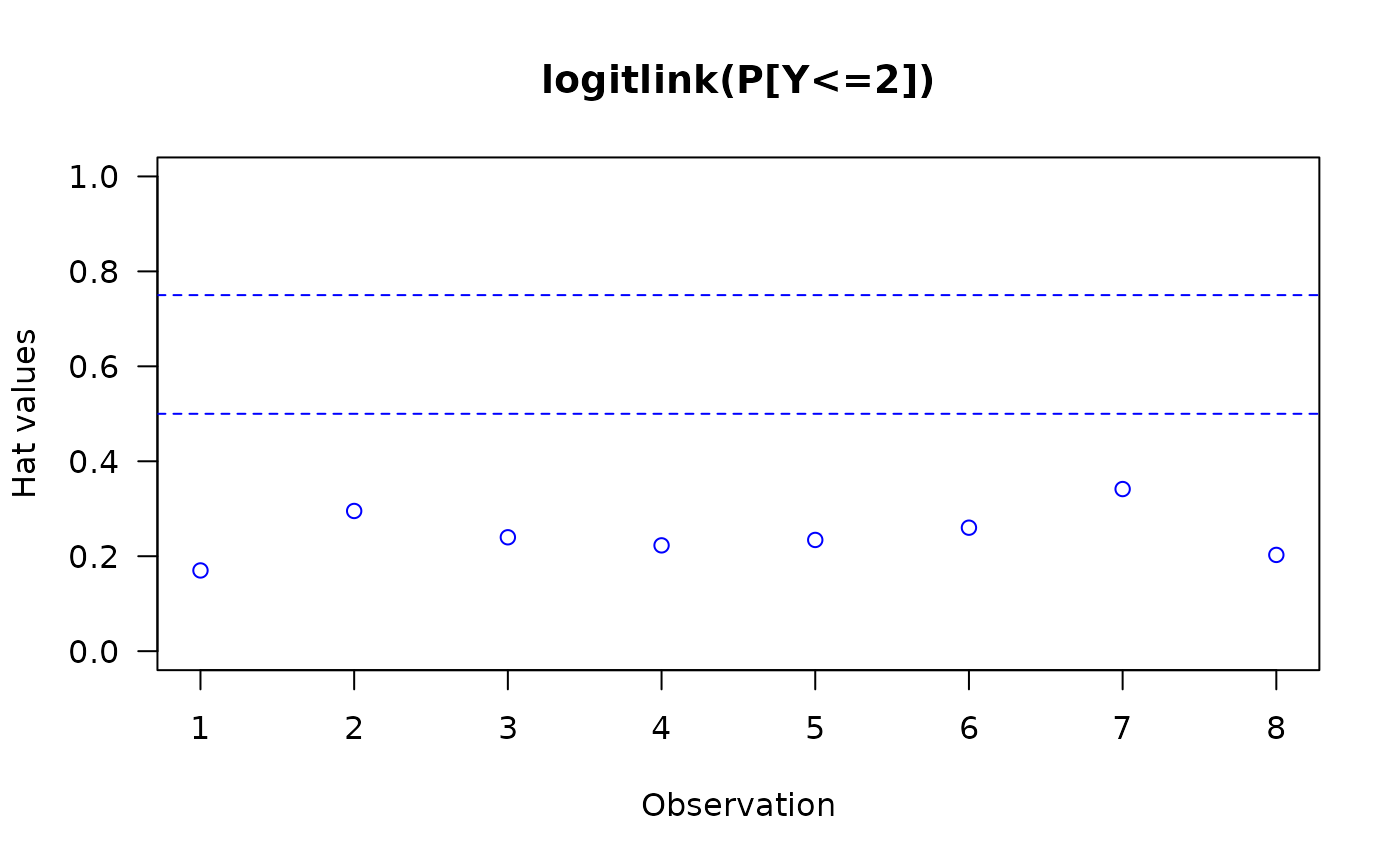
The invocation hatplot(vglmObject) should plot
the diagonal of the hat matrix for each of the \(M\)
linear/additive predictors.
By default, two horizontal dashed lines are added;
hat values higher than these ought to be checked.
Note
It is hoped, soon, that the full suite of functions described at
influence.measures will be written for VGLMs.
This will enable general regression deletion diagnostics to be
available for the entire VGLM class.
See also
Examples
# Proportional odds model, p.179, in McCullagh and Nelder (1989)
pneumo <- transform(pneumo, let = log(exposure.time))
fit <- vglm(cbind(normal, mild, severe) ~ let, cumulative, data = pneumo)
hatvalues(fit) # n x M matrix, with positive values
#> logitlink(P[Y<=1]) logitlink(P[Y<=2])
#> 1 0.2569868 0.1700224
#> 2 0.3424524 0.2952981
#> 3 0.2386174 0.2398881
#> 4 0.2154987 0.2228574
#> 5 0.2480763 0.2342776
#> 6 0.2668986 0.2601074
#> 7 0.3034072 0.3414842
#> 8 0.1613736 0.2027538
#> attr(,"predictors.names")
#> [1] "logitlink(P[Y<=1])" "logitlink(P[Y<=2])"
#> attr(,"ncol.X.vlm")
#> [1] 4
all.equal(sum(hatvalues(fit)), fit@rank) # Should be TRUE
#> [1] TRUE
if (FALSE) par(mfrow = c(1, 2))
hatplot(fit, ylim = c(0, 1), las = 1, col = "blue") # \dontrun{}