The Poisson Lognormal Distribution
polonoUC.RdDensity, distribution function and random generation for the Poisson lognormal distribution.
dpolono(x, meanlog = 0, sdlog = 1, bigx = 170, ...)
ppolono(q, meanlog = 0, sdlog = 1,
isOne = 1 - sqrt( .Machine$double.eps ), ...)
rpolono(n, meanlog = 0, sdlog = 1)Arguments
- n
number of observations. If
length(n) > 1then the length is taken to be the number required.- meanlog, sdlog
the mean and standard deviation of the normal distribution (on the log scale). They match the arguments in
Lognormal.- bigx
Numeric. This argument is for handling large values of
xand/or whenintegratefails. A first order Taylor series approximation [Equation (7) of Bulmer (1974)] is used at values ofxthat are greater or equal to this argument. Forbigx = 10, he showed that the approximation has a relative error less than 0.001 for values ofmeanlogandsdlog“likely to be encountered in practice”. The argument can be assignedInfin which case the approximation is not used.- isOne
Used to test whether the cumulative probabilities have effectively reached unity.
- ...
Arguments passed into
integrate.
Value
dpolono gives the density,
ppolono gives the distribution function, and
rpolono generates random deviates.
References
Bulmer, M. G. (1974). On fitting the Poisson lognormal distribution to species-abundance data. Biometrics, 30, 101–110.
Details
The Poisson lognormal distribution is similar to the negative binomial in that it can be motivated by a Poisson distribution whose mean parameter comes from a right skewed distribution (gamma for the negative binomial and lognormal for the Poisson lognormal distribution).
Note
By default,
dpolono involves numerical integration that is performed using
integrate. Consequently, computations are very
slow and numerical problems may occur
(if so then the use of ... may be needed).
Alternatively, for extreme values of x, meanlog,
sdlog, etc., the use of bigx = Inf avoids the call to
integrate, however the answer may be a little
inaccurate.
For the maximum likelihood estimation of the 2 parameters a VGAM
family function called polono(), say, has not been written yet.
See also
Examples
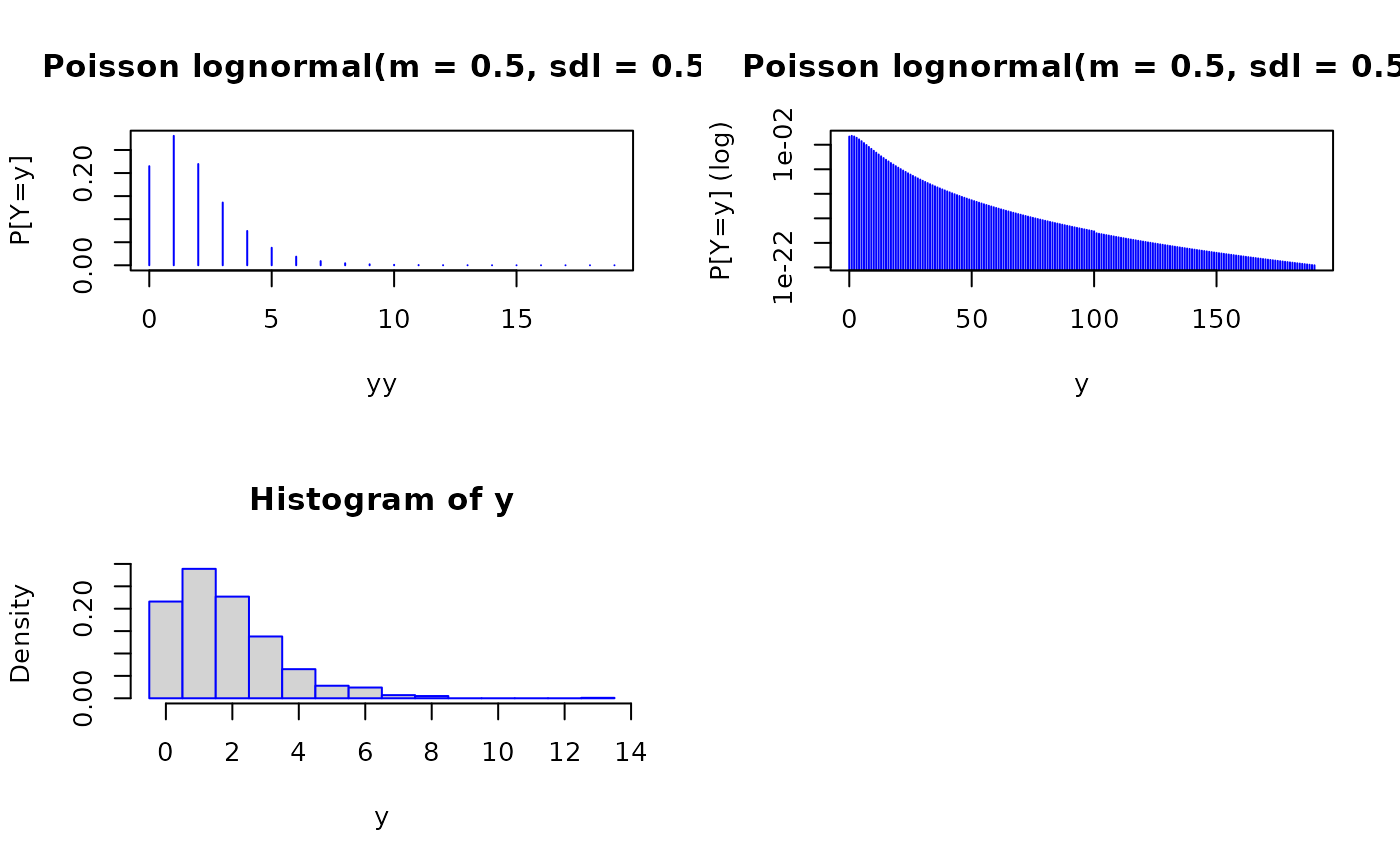
meanlog <- 0.5; sdlog <- 0.5; yy <- 0:19
sum(proby <- dpolono(yy, m = meanlog, sd = sdlog)) # Should be 1
#> [1] 0.9999955
max(abs(cumsum(proby) - ppolono(yy, m = meanlog, sd = sdlog))) # 0?
#> [1] 2.220446e-16
if (FALSE) opar = par(no.readonly = TRUE)
par(mfrow = c(2, 2))
plot(yy, proby, type = "h", col = "blue", ylab = "P[Y=y]", log = "",
main = paste0("Poisson lognormal(m = ", meanlog,
", sdl = ", sdlog, ")"))
y <- 0:190 # More extreme values; use the approxn & plot on a log scale
(sum(proby <- dpolono(y, m = meanlog, sd = sdlog, bigx = 100))) # 1?
#> [1] 1
plot(y, proby, type = "h", col = "blue", ylab = "P[Y=y] (log)", log = "y",
main = paste0("Poisson lognormal(m = ", meanlog,
", sdl = ", sdlog, ")")) # Note the kink at bigx
# Random number generation
table(y <- rpolono(n = 1000, m = meanlog, sd = sdlog))
#>
#> 0 1 2 3 4 5 6 7 8 13
#> 216 289 227 138 65 28 24 7 5 1
hist(y, breaks = ((-1):max(y))+0.5, prob = TRUE, border = "blue")
par(opar) # \dontrun{}
#> Error: object 'opar' not found