Zero-Altered Negative Binomial Distribution
zanegbinUC.RdDensity, distribution function, quantile function and random
generation for the zero-altered negative binomial distribution
with parameter pobs0.
dzanegbin(x, size, munb, pobs0 = 0, log = FALSE)
pzanegbin(q, size, munb, pobs0 = 0)
qzanegbin(p, size, munb, pobs0 = 0)
rzanegbin(n, size, munb, pobs0 = 0)Arguments
- x, q
vector of quantiles.
- p
vector of probabilities.
- n
number of observations. If
length(n) > 1then the length is taken to be the number required.- size, munb, log
Parameters from the ordinary negative binomial distribution (see
dnbinom). Some arguments have been renamed slightly.- pobs0
Probability of zero, called \(pobs0\). The default value of
pobs0 = 0corresponds to the response having a positive negative binomial distribution.
Details
The probability function of \(Y\) is 0 with
probability pobs0, else a positive negative
binomial(\(\mu_{nb}\), size) distribution.
Value
dzanegbin gives the density and
pzanegbin gives the distribution function,
qzanegbin gives the quantile function, and
rzanegbin generates random deviates.
Note
The argument pobs0 is recycled to the required length,
and must have values which lie in the interval \([0,1]\).
See also
Examples
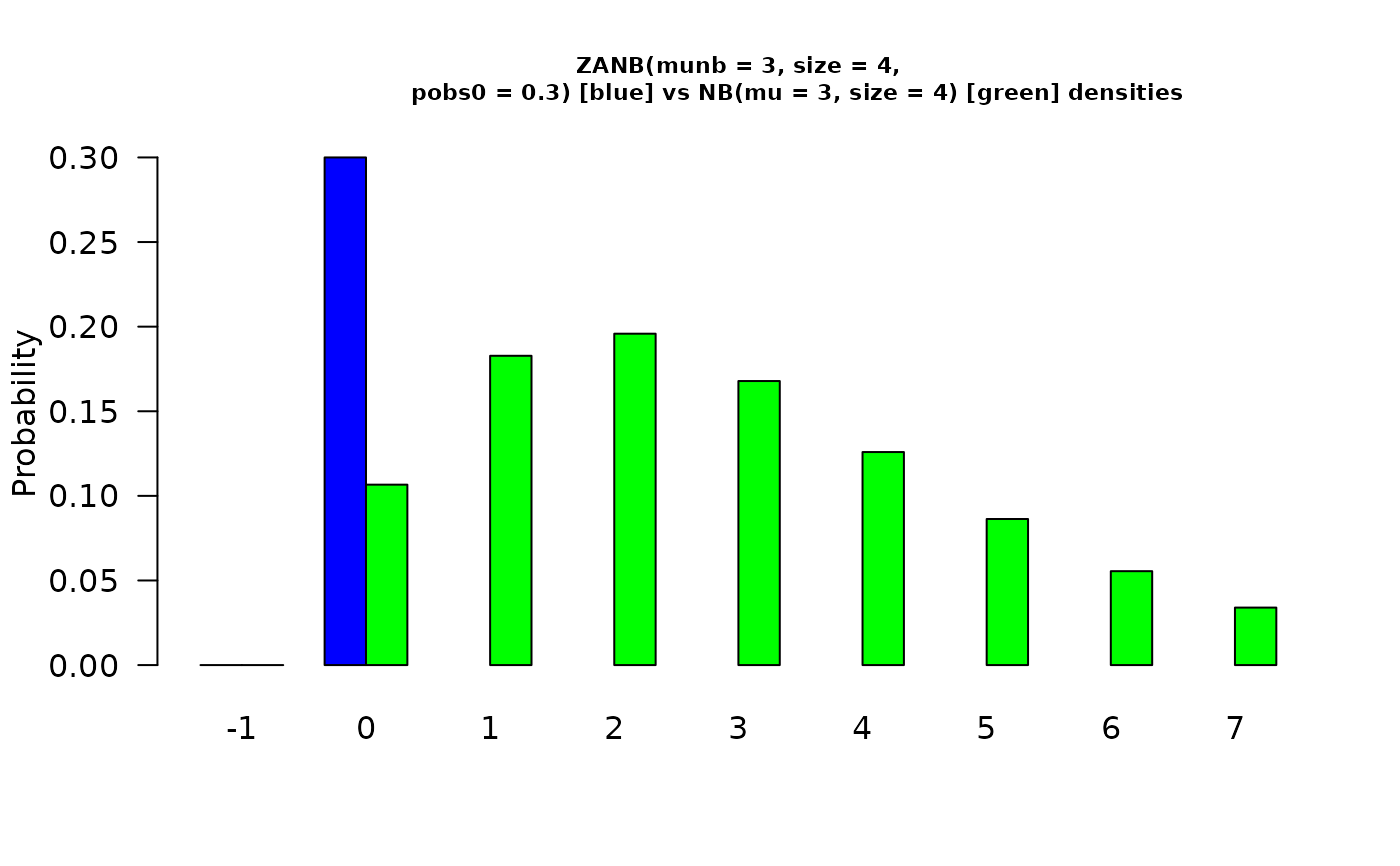
munb <- 3; size <- 4; pobs0 <- 0.3; x <- (-1):7
dzanegbin(x, munb = munb, size = size, pobs0 = pobs0)
#> [1] 0.0 0.3 NA NA NA NA NA NA NA
table(rzanegbin(100, munb = munb, size = size, pobs0 = pobs0))
#>
#> 0 1 2 3 4 5 6 7 9 10 11
#> 31 14 14 14 11 8 2 2 2 1 1
if (FALSE) x <- 0:10
barplot(rbind(dzanegbin(x, munb = munb, size = size, pobs0 = pobs0),
dnbinom(x, mu = munb, size = size)),
beside = TRUE, col = c("blue", "green"), cex.main = 0.7,
ylab = "Probability", names.arg = as.character(x), las = 1,
main = paste0("ZANB(munb = ", munb, ", size = ", size,",
pobs0 = ", pobs0,
") [blue] vs", " NB(mu = ", munb, ", size = ", size,
") [green] densities")) # \dontrun{}