Performs a simple test to check whether the prior is informative to the posterior. This idea, and the accompanying heuristics, were discussed in Gelman et al. 2017.
check_prior(model, method = "gelman", simulate_priors = TRUE, ...)
# S3 method for class 'brmsfit'
check_prior(
model,
method = "gelman",
simulate_priors = TRUE,
effects = "fixed",
component = "conditional",
parameters = NULL,
verbose = TRUE,
...
)Arguments
- model
A
stanreg,stanfit,brmsfit,blavaan, orMCMCglmmobject.- method
Can be
"gelman"or"lakeland". For the"gelman"method, if the SD of the posterior is more than 0.1 times the SD of the prior, then the prior is considered as informative. For the"lakeland"method, the prior is considered as informative if the posterior falls within the95%HDI of the prior.- simulate_priors
Should prior distributions be simulated using
simulate_prior()(default; faster) or sampled viaunupdate()(slower, more accurate).- ...
Currently not used.
- effects
Should variables for fixed effects (
"fixed"), random effects ("random") or both ("all") be returned? Only applies to mixed models. May be abbreviated.For models of from packages brms or rstanarm there are additional options:
"fixed"returns fixed effects."random_variance"return random effects parameters (variance and correlation components, e.g. those parameters that start withsd_orcor_)."grouplevel"returns random effects group level estimates, i.e. those parameters that start withr_."random"returns both"random_variance"and"grouplevel"."all"returns fixed effects and random effects variances."full"returns all parameters.
- component
Which type of parameters to return, such as parameters for the conditional model, the zero-inflated part of the model, the dispersion term, etc. See details in section Model Components. May be abbreviated. Note that the conditional component also refers to the count or mean component - names may differ, depending on the modeling package. There are three convenient shortcuts (not applicable to all model classes):
component = "all"returns all possible parameters.If
component = "location", location parameters such asconditional,zero_inflated,smooth_terms, orinstrumentsare returned (everything that are fixed or random effects - depending on theeffectsargument - but no auxiliary parameters).For
component = "distributional"(or"auxiliary"), components likesigma,dispersion,betaorprecision(and other auxiliary parameters) are returned.
- parameters
Regular expression pattern that describes the parameters that should be returned. Meta-parameters (like
lp__orprior_) are filtered by default, so only parameters that typically appear in thesummary()are returned. Useparametersto select specific parameters for the output.- verbose
Toggle off warnings.
Value
A data frame with two columns: The parameter names and the quality
of the prior (which might be "informative", "uninformative")
or "not determinable" if the prior distribution could not be
determined).
References
Gelman, A., Simpson, D., and Betancourt, M. (2017). The Prior Can Often Only Be Understood in the Context of the Likelihood. Entropy, 19(10), 555. doi:10.3390/e19100555
Examples
# \donttest{
library(bayestestR)
model <- rstanarm::stan_glm(mpg ~ wt + am, data = mtcars, chains = 1, refresh = 0)
check_prior(model, method = "gelman")
#> Parameter Prior_Quality
#> 1 (Intercept) informative
#> 2 wt uninformative
#> 3 am uninformative
check_prior(model, method = "lakeland")
#> Parameter Prior_Quality
#> 1 (Intercept) informative
#> 2 wt informative
#> 3 am informative
# An extreme example where both methods diverge:
model <- rstanarm::stan_glm(mpg ~ wt,
data = mtcars[1:3, ],
prior = normal(-3.3, 1, FALSE),
prior_intercept = normal(0, 1000, FALSE),
refresh = 0
)
check_prior(model, method = "gelman")
#> Parameter Prior_Quality
#> 1 (Intercept) uninformative
#> 2 wt informative
check_prior(model, method = "lakeland")
#> Parameter Prior_Quality
#> 1 (Intercept) informative
#> 2 wt misinformative
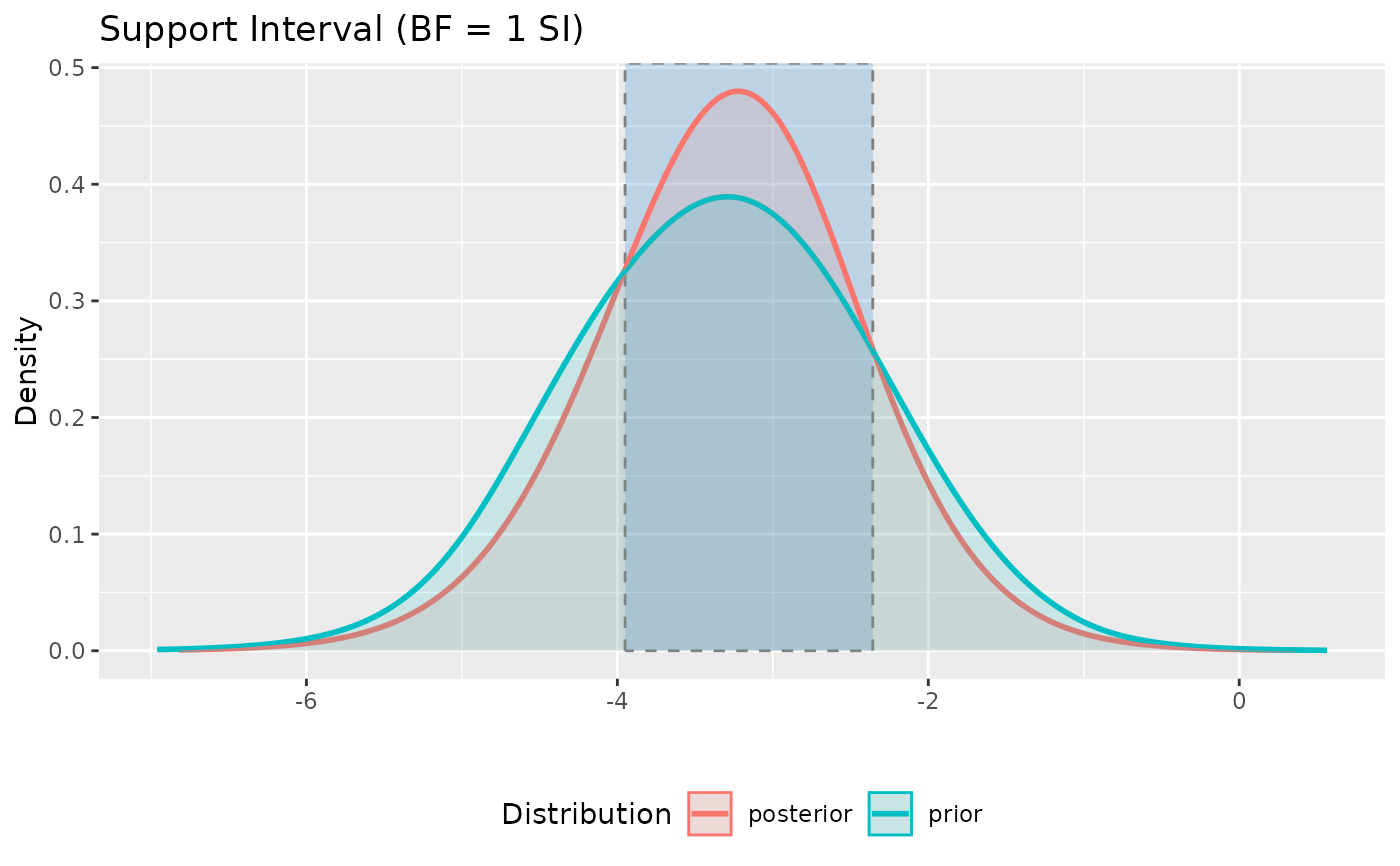
# can provide visual confirmation to the Lakeland method
plot(si(model, verbose = FALSE))
 # }
# }