Mean of a Moving Window
runmean.RdMoving (aka running, rolling) Window Mean calculated over a vector
Arguments
- x
numeric vector of length n or matrix with n rows. If
xis a matrix than each column will be processed separately.- k
width of moving window; must be an integer between 1 and n
- alg
an option to choose different algorithms
"C"- a version is written in C. It can handle non-finite numbers like NaN's and Inf's (likemean(x, na.rm = TRUE)). It works the fastest forendrule="mean"."fast"- second, even faster, C version. This algorithm does not work with non-finite numbers. It also works the fastest forendruleother than"mean"."R"- much slower code written in R. Useful for debugging and as documentation."exact"- same as"C", except that all additions are performed using algorithm which tracks and corrects addition round-off errors
- endrule
character string indicating how the values at the beginning and the end, of the data, should be treated. Only first and last
k2values at both ends are affected, wherek2is the half-bandwidthk2 = k %/% 2."mean"- applies the underlying function to smaller and smaller sections of the array. Equivalent to:for(i in 1:k2) out[i] = mean(x[1:(i+k2)]). This option is implemented in C ifalg="C", otherwise is done in R."trim"- trim the ends; output array length is equal tolength(x)-2*k2 (out = out[(k2+1):(n-k2)]). This option mimics output ofapply(embed(x,k),1,mean)and other related functions."keep"- fill the ends with numbers fromxvector(out[1:k2] = x[1:k2])"constant"- fill the ends with first and last calculated value in output array(out[1:k2] = out[k2+1])"NA"- fill the ends with NA's(out[1:k2] = NA)"func"- same as"mean"but implimented in R. This option could be very slow, and is included mostly for testing
Similar to
endruleinrunmedfunction which has the following options: “c("median", "keep", "constant")” .- align
specifies whether result should be centered (default), left-aligned or right-aligned. If
endrule="mean" then settingalignto "left" or "right" will fall back on slower implementation equivalent toendrule="func".
Details
Apart from the end values, the result of y = runmean(x, k) is the same as
“for(j=(1+k2):(n-k2)) y[j]=mean(x[(j-k2):(j+k2)])”.
The main incentive to write this set of functions was relative slowness of
majority of moving window functions available in R and its packages. With the
exception of runmed, a running window median function, all
functions listed in "see also" section are slower than very inefficient
“apply(embed(x,k),1,FUN)” approach. Relative
speed of runmean function is O(n).
Function EndRule applies one of the five methods (see endrule
argument) to process end-points of the input array x. In current
version of the code the default endrule="mean" option is calculated
within C code. That is done to improve speed in case of large moving windows.
In case of runmean(..., alg="exact") function a special algorithm is
used (see references section) to ensure that round-off errors do not
accumulate. As a result runmean is more accurate than
filter(x, rep(1/k,k)) and runmean(..., alg="C")
functions.
Value
Returns a numeric vector or matrix of the same size as x. Only in case of
endrule="trim" the output vectors will be shorter and output matrices
will have fewer rows.
References
About round-off error correction used in
runmean: Shewchuk, Jonathan Adaptive Precision Floating-Point Arithmetic and Fast Robust Geometric Predicates, http://www-2.cs.cmu.edu/afs/cs/project/quake/public/papers/robust-arithmetic.ps
Note
Function runmean(..., alg="exact") is based by code by Vadim Ogranovich,
which is based on Python code (see last reference), pointed out by Gabor
Grothendieck.
See also
Links related to:
moving mean -
mean,kernapply,filter,decompose,stl,rollmeanfrom zoo library,subsumsfrom magic library,Other moving window functions from this package:
runmin,runmax,runquantile,runmadandrunsdgeneric running window functions:
apply(embed(x,k), 1, FUN)(fastest),runningfrom gtools package (extremely slow for this purpose),subsumsfrom magic library can perform running window operations on data with any dimensions.
Examples
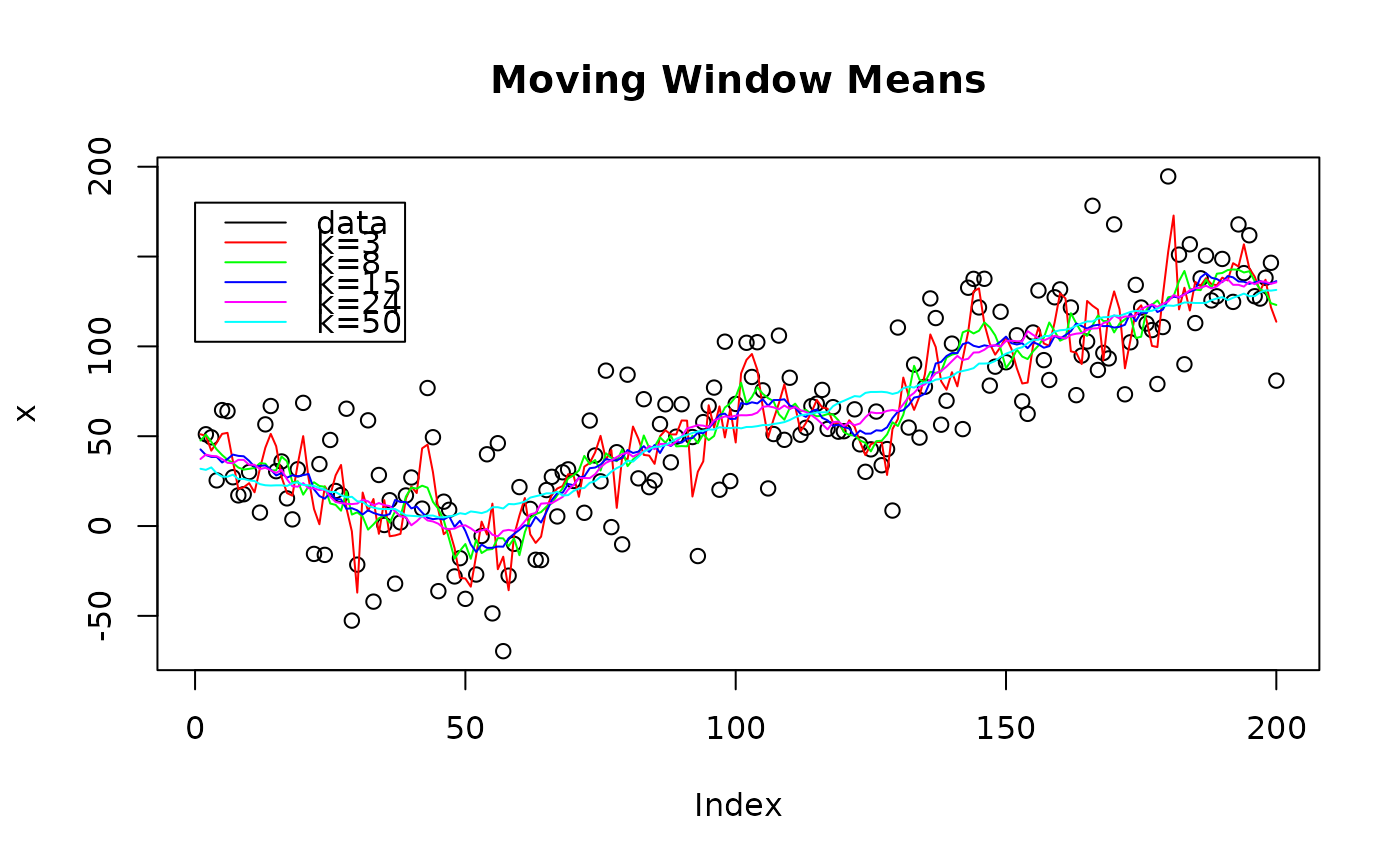
# show runmean for different window sizes
n=200;
x = rnorm(n,sd=30) + abs(seq(n)-n/4)
x[seq(1,n,10)] = NaN; # add NANs
col = c("black", "red", "green", "blue", "magenta", "cyan")
plot(x, col=col[1], main = "Moving Window Means")
lines(runmean(x, 3), col=col[2])
lines(runmean(x, 8), col=col[3])
lines(runmean(x,15), col=col[4])
lines(runmean(x,24), col=col[5])
lines(runmean(x,50), col=col[6])
lab = c("data", "k=3", "k=8", "k=15", "k=24", "k=50")
legend(0,0.9*n, lab, col=col, lty=1 )
 # basic tests against 2 standard R approaches
k=25; n=200;
x = rnorm(n,sd=30) + abs(seq(n)-n/4) # create random data
a = runmean(x,k, endrule="trim") # tested function
b = apply(embed(x,k), 1, mean) # approach #1
c = cumsum(c( sum(x[1:k]), diff(x,k) ))/k # approach #2
eps = .Machine$double.eps ^ 0.5
stopifnot(all(abs(a-b)<eps));
stopifnot(all(abs(a-c)<eps));
# test against loop approach
# this test works fine at the R prompt but fails during package check - need to investigate
k=25;
data(iris)
x = iris[,1]
n = length(x)
x[seq(1,n,11)] = NaN; # add NANs
k2 = k
k1 = k-k2-1
a = runmean(x, k)
b = array(0,n)
for(j in 1:n) {
lo = max(1, j-k1)
hi = min(n, j+k2)
b[j] = mean(x[lo:hi], na.rm = TRUE)
}
#stopifnot(all(abs(a-b)<eps)); # commented out for time beeing - on to do list
# compare calculation at array ends
a = runmean(x, k, endrule="mean") # fast C code
b = runmean(x, k, endrule="func") # slow R code
stopifnot(all(abs(a-b)<eps));
# Testing of different methods to each other for non-finite data
# Only alg "C" and "exact" can handle not finite numbers
eps = .Machine$double.eps ^ 0.5
n=200; k=51;
x = rnorm(n,sd=30) + abs(seq(n)-n/4) # nice behaving data
x[seq(1,n,10)] = NaN; # add NANs
x[seq(1,n, 9)] = Inf; # add infinities
b = runmean( x, k, alg="C")
c = runmean( x, k, alg="exact")
stopifnot(all(abs(b-c)<eps));
# Test if moving windows forward and backward gives the same results
# Test also performed on data with non-finite numbers
a = runmean(x , alg="C", k)
b = runmean(x[n:1], alg="C", k)
stopifnot(all(abs(a[n:1]-b)<eps));
a = runmean(x , alg="exact", k)
b = runmean(x[n:1], alg="exact", k)
stopifnot(all(abs(a[n:1]-b)<eps));
# test vector vs. matrix inputs, especially for the edge handling
nRow=200; k=25; nCol=10
x = rnorm(nRow,sd=30) + abs(seq(nRow)-n/4)
x[seq(1,nRow,10)] = NaN; # add NANs
X = matrix(rep(x, nCol ), nRow, nCol) # replicate x in columns of X
a = runmean(x, k)
b = runmean(X, k)
stopifnot(all(abs(a-b[,1])<eps)); # vector vs. 2D array
stopifnot(all(abs(b[,1]-b[,nCol])<eps)); # compare rows within 2D array
# Exhaustive testing of different methods to each other for different windows
numeric.test = function (x, k) {
a = runmean( x, k, alg="fast")
b = runmean( x, k, alg="C")
c = runmean( x, k, alg="exact")
d = runmean( x, k, alg="R", endrule="func")
eps = .Machine$double.eps ^ 0.5
stopifnot(all(abs(a-b)<eps));
stopifnot(all(abs(b-c)<eps));
stopifnot(all(abs(c-d)<eps));
}
n=200;
x = rnorm(n,sd=30) + abs(seq(n)-n/4) # nice behaving data
for(i in 1:5) numeric.test(x, i) # test small window sizes
for(i in 1:5) numeric.test(x, n-i+1) # test large window size
# speed comparison
if (FALSE) { # \dontrun{
x=runif(1e7); k=1e4;
system.time(runmean(x,k,alg="fast"))
system.time(runmean(x,k,alg="C"))
system.time(runmean(x,k,alg="exact"))
system.time(runmean(x,k,alg="R")) # R version of the function
x=runif(1e5); k=1e2; # reduce vector and window sizes
system.time(runmean(x,k,alg="R")) # R version of the function
system.time(apply(embed(x,k), 1, mean)) # standard R approach
system.time(filter(x, rep(1/k,k), sides=2)) # the fastest alternative I know
} # }
# show different runmean algorithms with data spanning many orders of magnitude
n=30; k=5;
x = rep(100/3,n)
d=1e10
x[5] = d;
x[13] = d;
x[14] = d*d;
x[15] = d*d*d;
x[16] = d*d*d*d;
x[17] = d*d*d*d*d;
a = runmean(x, k, alg="fast" )
b = runmean(x, k, alg="C" )
c = runmean(x, k, alg="exact")
y = t(rbind(x,a,b,c))
y
#> x a b c
#> [1,] 3.333333e+01 3.333333e+01 3.333333e+01 3.333333e+01
#> [2,] 3.333333e+01 3.333333e+01 3.333333e+01 3.333333e+01
#> [3,] 3.333333e+01 2.000000e+09 2.000000e+09 2.000000e+09
#> [4,] 3.333333e+01 2.000000e+09 2.000000e+09 2.000000e+09
#> [5,] 1.000000e+10 2.000000e+09 2.000000e+09 2.000000e+09
#> [6,] 3.333333e+01 2.000000e+09 2.000000e+09 2.000000e+09
#> [7,] 3.333333e+01 2.000000e+09 2.000000e+09 2.000000e+09
#> [8,] 3.333333e+01 3.333333e+01 3.333333e+01 3.333333e+01
#> [9,] 3.333333e+01 3.333333e+01 3.333333e+01 3.333333e+01
#> [10,] 3.333333e+01 3.333333e+01 3.333333e+01 3.333333e+01
#> [11,] 3.333333e+01 2.000000e+09 2.000000e+09 2.000000e+09
#> [12,] 3.333333e+01 2.000000e+19 2.000000e+19 2.000000e+19
#> [13,] 1.000000e+10 2.000000e+29 2.000000e+29 2.000000e+29
#> [14,] 1.000000e+20 2.000000e+39 2.000000e+39 2.000000e+39
#> [15,] 1.000000e+30 2.000000e+49 2.000000e+49 2.000000e+49
#> [16,] 1.000000e+40 2.000000e+49 2.000000e+49 2.000000e+49
#> [17,] 1.000000e+50 2.000000e+49 2.000000e+49 2.000000e+49
#> [18,] 3.333333e+01 2.000000e+49 2.000000e+49 2.000000e+49
#> [19,] 3.333333e+01 2.000000e+49 2.000000e+49 2.000000e+49
#> [20,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [21,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [22,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [23,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [24,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [25,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [26,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [27,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [28,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [29,] 3.333333e+01 -8.333333e+00 -8.333333e+00 3.333333e+01
#> [30,] 3.333333e+01 -2.222222e+01 -2.222222e+01 3.333333e+01
# basic tests against 2 standard R approaches
k=25; n=200;
x = rnorm(n,sd=30) + abs(seq(n)-n/4) # create random data
a = runmean(x,k, endrule="trim") # tested function
b = apply(embed(x,k), 1, mean) # approach #1
c = cumsum(c( sum(x[1:k]), diff(x,k) ))/k # approach #2
eps = .Machine$double.eps ^ 0.5
stopifnot(all(abs(a-b)<eps));
stopifnot(all(abs(a-c)<eps));
# test against loop approach
# this test works fine at the R prompt but fails during package check - need to investigate
k=25;
data(iris)
x = iris[,1]
n = length(x)
x[seq(1,n,11)] = NaN; # add NANs
k2 = k
k1 = k-k2-1
a = runmean(x, k)
b = array(0,n)
for(j in 1:n) {
lo = max(1, j-k1)
hi = min(n, j+k2)
b[j] = mean(x[lo:hi], na.rm = TRUE)
}
#stopifnot(all(abs(a-b)<eps)); # commented out for time beeing - on to do list
# compare calculation at array ends
a = runmean(x, k, endrule="mean") # fast C code
b = runmean(x, k, endrule="func") # slow R code
stopifnot(all(abs(a-b)<eps));
# Testing of different methods to each other for non-finite data
# Only alg "C" and "exact" can handle not finite numbers
eps = .Machine$double.eps ^ 0.5
n=200; k=51;
x = rnorm(n,sd=30) + abs(seq(n)-n/4) # nice behaving data
x[seq(1,n,10)] = NaN; # add NANs
x[seq(1,n, 9)] = Inf; # add infinities
b = runmean( x, k, alg="C")
c = runmean( x, k, alg="exact")
stopifnot(all(abs(b-c)<eps));
# Test if moving windows forward and backward gives the same results
# Test also performed on data with non-finite numbers
a = runmean(x , alg="C", k)
b = runmean(x[n:1], alg="C", k)
stopifnot(all(abs(a[n:1]-b)<eps));
a = runmean(x , alg="exact", k)
b = runmean(x[n:1], alg="exact", k)
stopifnot(all(abs(a[n:1]-b)<eps));
# test vector vs. matrix inputs, especially for the edge handling
nRow=200; k=25; nCol=10
x = rnorm(nRow,sd=30) + abs(seq(nRow)-n/4)
x[seq(1,nRow,10)] = NaN; # add NANs
X = matrix(rep(x, nCol ), nRow, nCol) # replicate x in columns of X
a = runmean(x, k)
b = runmean(X, k)
stopifnot(all(abs(a-b[,1])<eps)); # vector vs. 2D array
stopifnot(all(abs(b[,1]-b[,nCol])<eps)); # compare rows within 2D array
# Exhaustive testing of different methods to each other for different windows
numeric.test = function (x, k) {
a = runmean( x, k, alg="fast")
b = runmean( x, k, alg="C")
c = runmean( x, k, alg="exact")
d = runmean( x, k, alg="R", endrule="func")
eps = .Machine$double.eps ^ 0.5
stopifnot(all(abs(a-b)<eps));
stopifnot(all(abs(b-c)<eps));
stopifnot(all(abs(c-d)<eps));
}
n=200;
x = rnorm(n,sd=30) + abs(seq(n)-n/4) # nice behaving data
for(i in 1:5) numeric.test(x, i) # test small window sizes
for(i in 1:5) numeric.test(x, n-i+1) # test large window size
# speed comparison
if (FALSE) { # \dontrun{
x=runif(1e7); k=1e4;
system.time(runmean(x,k,alg="fast"))
system.time(runmean(x,k,alg="C"))
system.time(runmean(x,k,alg="exact"))
system.time(runmean(x,k,alg="R")) # R version of the function
x=runif(1e5); k=1e2; # reduce vector and window sizes
system.time(runmean(x,k,alg="R")) # R version of the function
system.time(apply(embed(x,k), 1, mean)) # standard R approach
system.time(filter(x, rep(1/k,k), sides=2)) # the fastest alternative I know
} # }
# show different runmean algorithms with data spanning many orders of magnitude
n=30; k=5;
x = rep(100/3,n)
d=1e10
x[5] = d;
x[13] = d;
x[14] = d*d;
x[15] = d*d*d;
x[16] = d*d*d*d;
x[17] = d*d*d*d*d;
a = runmean(x, k, alg="fast" )
b = runmean(x, k, alg="C" )
c = runmean(x, k, alg="exact")
y = t(rbind(x,a,b,c))
y
#> x a b c
#> [1,] 3.333333e+01 3.333333e+01 3.333333e+01 3.333333e+01
#> [2,] 3.333333e+01 3.333333e+01 3.333333e+01 3.333333e+01
#> [3,] 3.333333e+01 2.000000e+09 2.000000e+09 2.000000e+09
#> [4,] 3.333333e+01 2.000000e+09 2.000000e+09 2.000000e+09
#> [5,] 1.000000e+10 2.000000e+09 2.000000e+09 2.000000e+09
#> [6,] 3.333333e+01 2.000000e+09 2.000000e+09 2.000000e+09
#> [7,] 3.333333e+01 2.000000e+09 2.000000e+09 2.000000e+09
#> [8,] 3.333333e+01 3.333333e+01 3.333333e+01 3.333333e+01
#> [9,] 3.333333e+01 3.333333e+01 3.333333e+01 3.333333e+01
#> [10,] 3.333333e+01 3.333333e+01 3.333333e+01 3.333333e+01
#> [11,] 3.333333e+01 2.000000e+09 2.000000e+09 2.000000e+09
#> [12,] 3.333333e+01 2.000000e+19 2.000000e+19 2.000000e+19
#> [13,] 1.000000e+10 2.000000e+29 2.000000e+29 2.000000e+29
#> [14,] 1.000000e+20 2.000000e+39 2.000000e+39 2.000000e+39
#> [15,] 1.000000e+30 2.000000e+49 2.000000e+49 2.000000e+49
#> [16,] 1.000000e+40 2.000000e+49 2.000000e+49 2.000000e+49
#> [17,] 1.000000e+50 2.000000e+49 2.000000e+49 2.000000e+49
#> [18,] 3.333333e+01 2.000000e+49 2.000000e+49 2.000000e+49
#> [19,] 3.333333e+01 2.000000e+49 2.000000e+49 2.000000e+49
#> [20,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [21,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [22,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [23,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [24,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [25,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [26,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [27,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [28,] 3.333333e+01 0.000000e+00 0.000000e+00 3.333333e+01
#> [29,] 3.333333e+01 -8.333333e+00 -8.333333e+00 3.333333e+01
#> [30,] 3.333333e+01 -2.222222e+01 -2.222222e+01 3.333333e+01