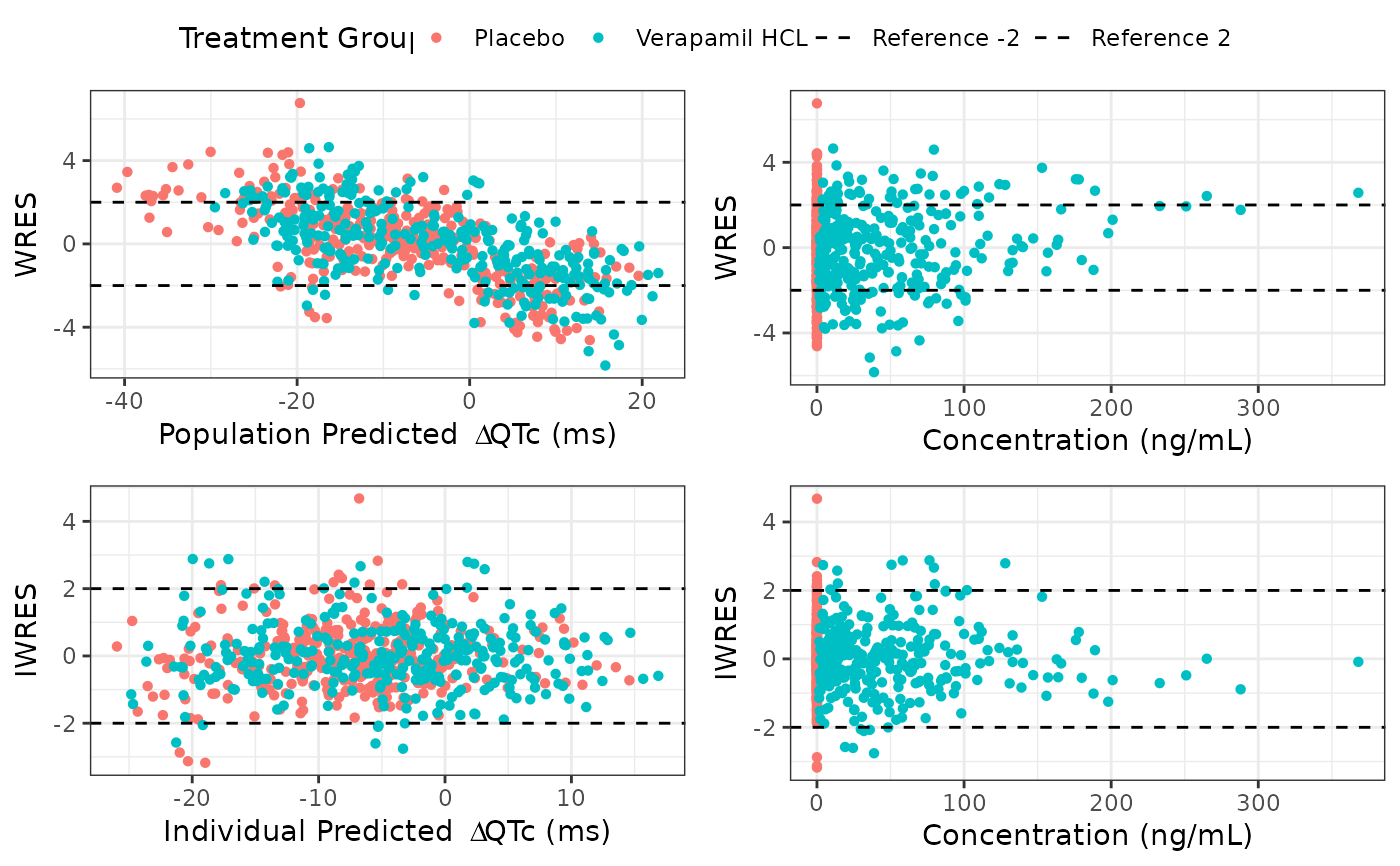
Plots residuals vs predicted dQTCF and concentration
gof_residuals_plots.RdPlots residuals vs predicted dQTCF and concentration
Arguments
- data
A data frame containing C-QT analysis dataset
- fit
An nlme::lme model object from model fitting
- dv_col
An unquoted column name for dependent variable measurements
- conc_col
An unquoted column name for drug concentration measurements
- ntime_col
An unquoted column name for nominal time since dose
- trt_col
An unquoted column name for treatment group"
- conc_xlabel
A string of concentration xlabel
- dv_label
A string of dv label (default: bquote(Delta ~ 'QTc (ms)'))
- residual_references
Numeric vector of reference residual lines to add, default -2 and 2
- legend_location
String for legend position (top, bottom, left, right)
- style
A named list of arguments passed to style_plot()
Value
a plot
Examples
data_proc <- preprocess(cqtkit_data_verapamil)
fit <- fit_prespecified_model(
data_proc,
deltaQTCF,
ID,
CONC,
deltaQTCFBL,
TRTG,
TAFD,
"REML",
TRUE
)
gof_residuals_plots(
data_proc,
fit,
deltaQTCF,
CONC,
NTLD,
TRTG,
legend_location = "top")