FlexMix Clustering Demo Driver
FLXmclust.RdThese are demo drivers for flexmix implementing
model-based clustering of Gaussian data.
FLXMCmvnorm(formula = . ~ ., diagonal = TRUE)
FLXMCnorm1(formula = . ~ .)Arguments
- formula
A formula which is interpreted relative to the formula specified in the call to
flexmixusingupdate.formula. Only the left-hand side (response) of the formula is used. Default is to use the originalflexmixmodel formula.- diagonal
If
TRUE, then the covariance matrix of the components is restricted to diagonal matrices.
Details
This is mostly meant as a demo for FlexMix driver programming, you
should also look at package mclust for real
applications. FLXMCmvnorm clusters multivariate data,
FLXMCnorm1 univariate data. In the latter case smart
initialization is important, see the example below.
Value
FLXMCmvnorm returns an object of class FLXMC.
References
Friedrich Leisch. FlexMix: A general framework for finite mixture models and latent class regression in R. Journal of Statistical Software, 11(8), 2004. doi:10.18637/jss.v011.i08
See also
Examples
data("Nclus", package = "flexmix")
require("MASS")
eqscplot(Nclus)
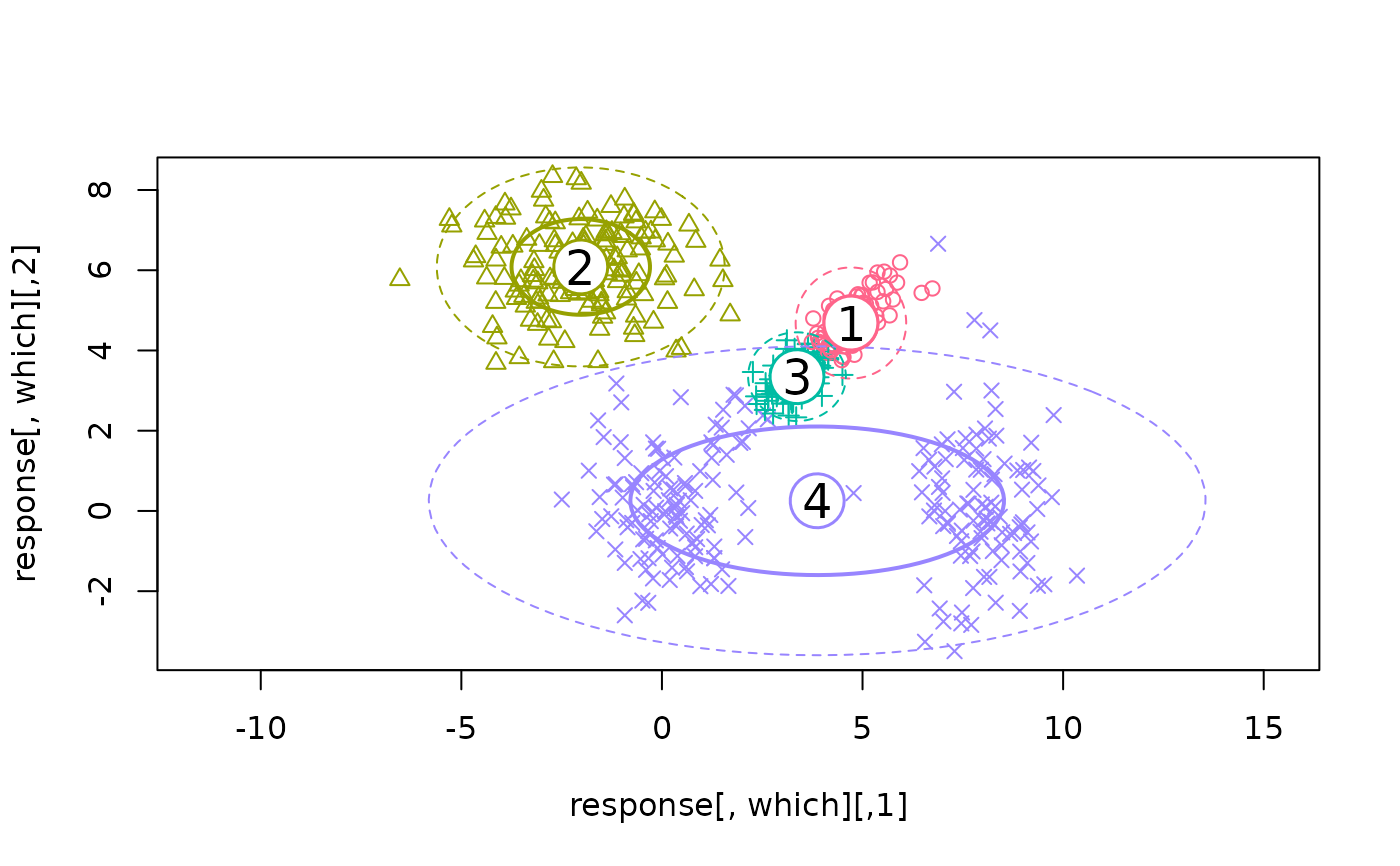
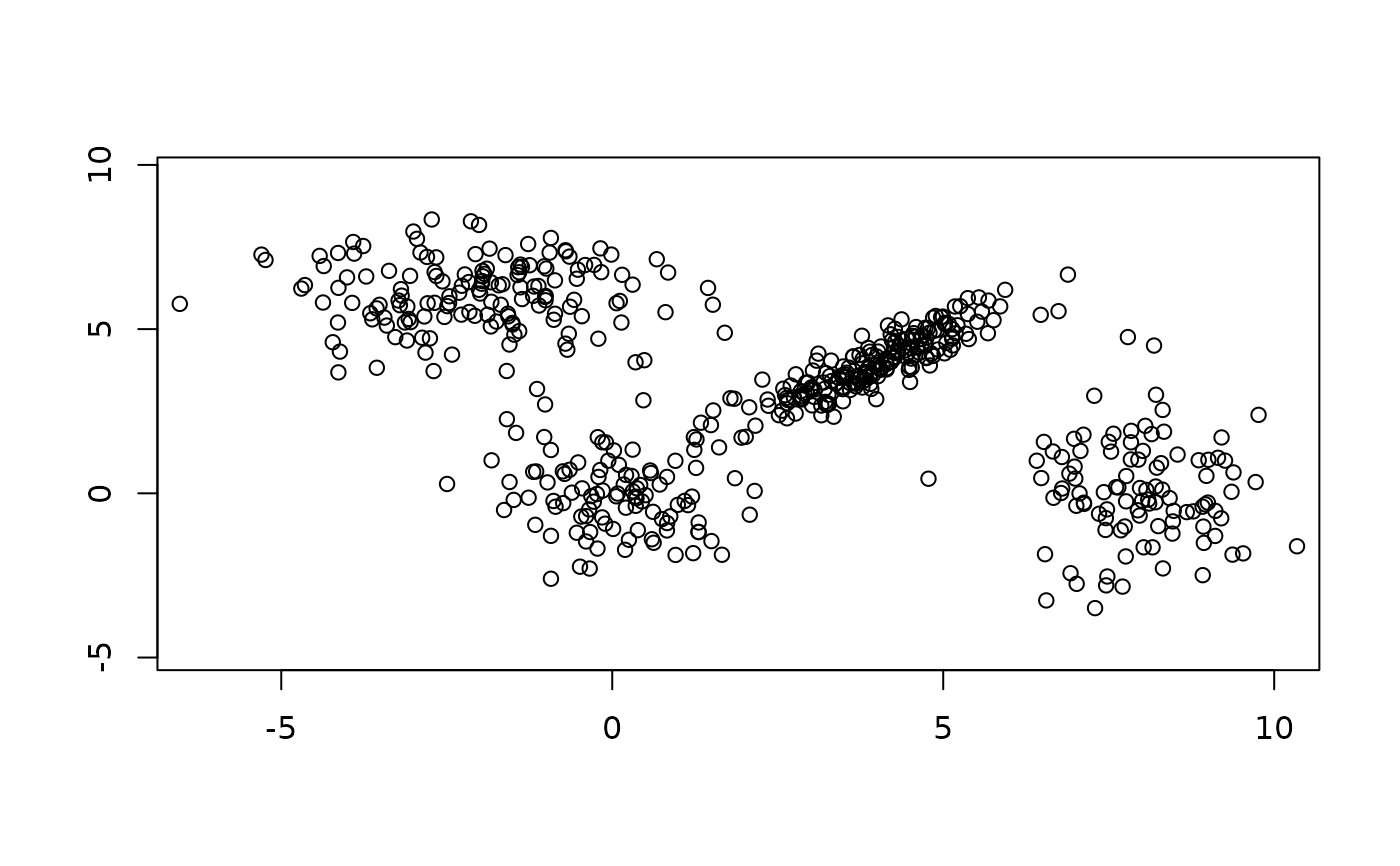 ## This model is wrong (one component has a non-diagonal cov matrix)
ex1 <- flexmix(Nclus ~ 1, k = 4, model = FLXMCmvnorm())
print(ex1)
#>
#> Call:
#> flexmix(formula = Nclus ~ 1, k = 4, model = FLXMCmvnorm())
#>
#> Cluster sizes:
#> 1 2 3 4
#> 96 149 92 213
#>
#> convergence after 194 iterations
plotEll(ex1, Nclus)
## This model is wrong (one component has a non-diagonal cov matrix)
ex1 <- flexmix(Nclus ~ 1, k = 4, model = FLXMCmvnorm())
print(ex1)
#>
#> Call:
#> flexmix(formula = Nclus ~ 1, k = 4, model = FLXMCmvnorm())
#>
#> Cluster sizes:
#> 1 2 3 4
#> 96 149 92 213
#>
#> convergence after 194 iterations
plotEll(ex1, Nclus)
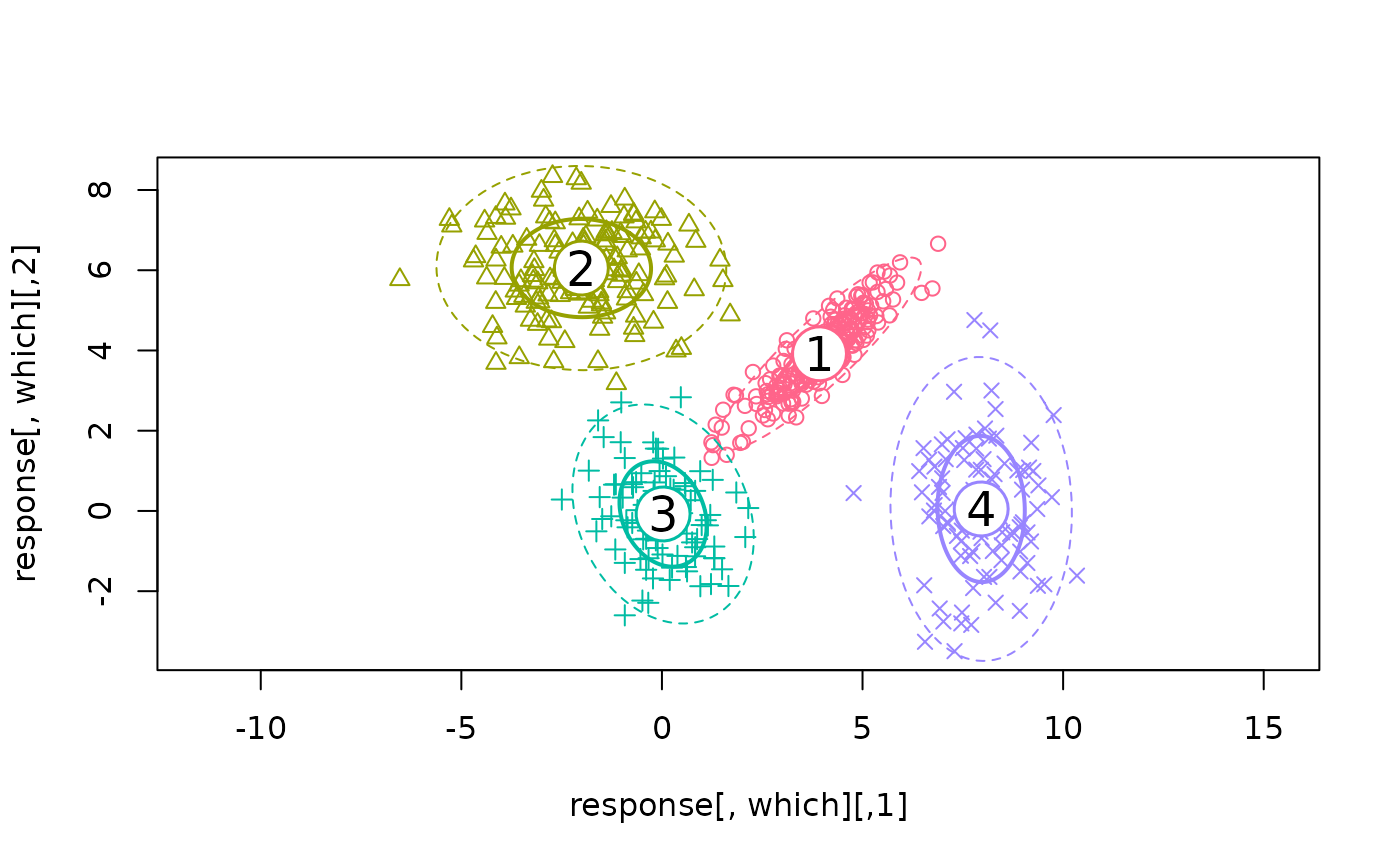
 ## True model, wrong number of components
ex2 <- flexmix(Nclus ~ 1, k = 6, model = FLXMCmvnorm(diagonal = FALSE))
print(ex2)
#>
#> Call:
#> flexmix(formula = Nclus ~ 1, k = 6, model = FLXMCmvnorm(diagonal = FALSE))
#>
#> Cluster sizes:
#> 1 2 3 4
#> 204 150 96 100
#>
#> convergence after 26 iterations
plotEll(ex2, Nclus)
## True model, wrong number of components
ex2 <- flexmix(Nclus ~ 1, k = 6, model = FLXMCmvnorm(diagonal = FALSE))
print(ex2)
#>
#> Call:
#> flexmix(formula = Nclus ~ 1, k = 6, model = FLXMCmvnorm(diagonal = FALSE))
#>
#> Cluster sizes:
#> 1 2 3 4
#> 204 150 96 100
#>
#> convergence after 26 iterations
plotEll(ex2, Nclus)
 ## Get parameters of first component
parameters(ex2, component = 1)
#> Comp.1
#> center1 3.9270274
#> center2 3.9177159
#> cov1 1.0737947
#> cov2 0.9109258
#> cov3 0.9109258
#> cov4 0.9604122
## Have a look at the posterior probabilies of 10 random observations
ok <- sample(1:nrow(Nclus), 10)
p <- posterior(ex2)[ok, ]
p
#> [,1] [,2] [,3] [,4]
#> [1,] 2.666664e-135 5.358805e-26 6.475614e-20 1.000000e+00
#> [2,] 3.459977e-07 2.271399e-11 9.999997e-01 1.900477e-15
#> [3,] 1.359455e-55 1.000000e+00 4.095438e-09 3.450189e-22
#> [4,] 1.714111e-88 3.551478e-20 1.179123e-21 1.000000e+00
#> [5,] 5.734686e-128 1.000000e+00 2.358708e-11 3.675333e-40
#> [6,] 5.154188e-08 1.768465e-12 9.999999e-01 9.534319e-18
#> [7,] 9.999906e-01 8.832400e-06 1.310348e-09 5.723562e-07
#> [8,] 9.999382e-01 6.020852e-05 6.204183e-12 1.635255e-06
#> [9,] 4.789248e-102 6.547787e-22 8.501703e-21 1.000000e+00
#> [10,] 9.999926e-01 6.017151e-06 2.752451e-09 1.418790e-06
## The following two should be the same
max.col(p)
#> [1] 4 3 2 4 2 3 1 1 4 1
clusters(ex2)[ok]
#> [1] 4 3 2 4 2 3 1 1 4 1
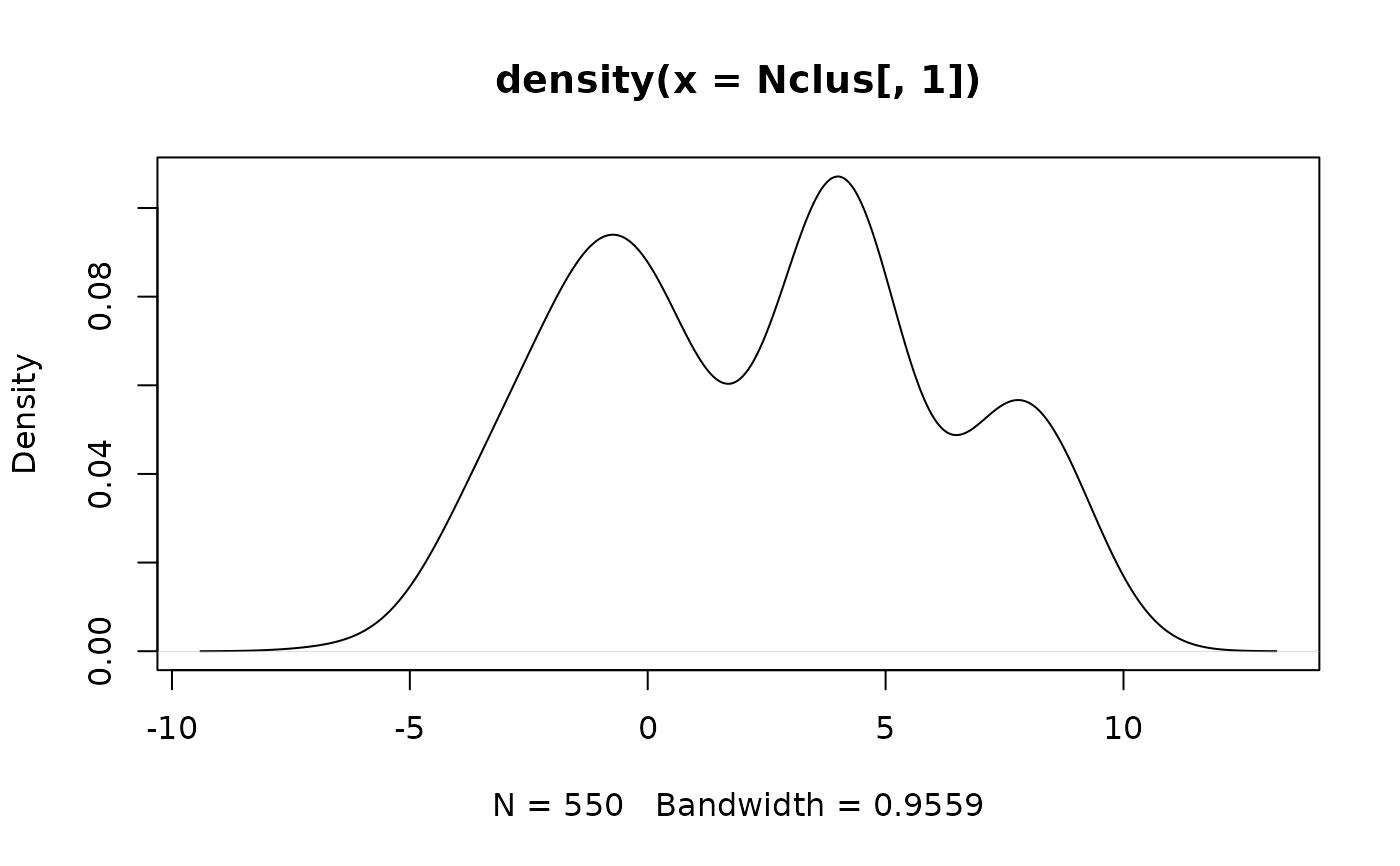
## Now try the univariate case
plot(density(Nclus[, 1]))
## Get parameters of first component
parameters(ex2, component = 1)
#> Comp.1
#> center1 3.9270274
#> center2 3.9177159
#> cov1 1.0737947
#> cov2 0.9109258
#> cov3 0.9109258
#> cov4 0.9604122
## Have a look at the posterior probabilies of 10 random observations
ok <- sample(1:nrow(Nclus), 10)
p <- posterior(ex2)[ok, ]
p
#> [,1] [,2] [,3] [,4]
#> [1,] 2.666664e-135 5.358805e-26 6.475614e-20 1.000000e+00
#> [2,] 3.459977e-07 2.271399e-11 9.999997e-01 1.900477e-15
#> [3,] 1.359455e-55 1.000000e+00 4.095438e-09 3.450189e-22
#> [4,] 1.714111e-88 3.551478e-20 1.179123e-21 1.000000e+00
#> [5,] 5.734686e-128 1.000000e+00 2.358708e-11 3.675333e-40
#> [6,] 5.154188e-08 1.768465e-12 9.999999e-01 9.534319e-18
#> [7,] 9.999906e-01 8.832400e-06 1.310348e-09 5.723562e-07
#> [8,] 9.999382e-01 6.020852e-05 6.204183e-12 1.635255e-06
#> [9,] 4.789248e-102 6.547787e-22 8.501703e-21 1.000000e+00
#> [10,] 9.999926e-01 6.017151e-06 2.752451e-09 1.418790e-06
## The following two should be the same
max.col(p)
#> [1] 4 3 2 4 2 3 1 1 4 1
clusters(ex2)[ok]
#> [1] 4 3 2 4 2 3 1 1 4 1
## Now try the univariate case
plot(density(Nclus[, 1]))
 ex3 <- flexmix(Nclus[, 1] ~ 1, cluster = cut(Nclus[, 1], 3),
model = FLXMCnorm1())
ex3
#>
#> Call:
#> flexmix(formula = Nclus[, 1] ~ 1, cluster = cut(Nclus[, 1], 3),
#> model = FLXMCnorm1())
#>
#> Cluster sizes:
#> 1 2 3
#> 262 186 102
#>
#> convergence after 128 iterations
parameters(ex3)
#> Comp.1 Comp.2 Comp.3
#> mean -0.9217948 4.0600622 7.9189585
#> sd 1.8563637 0.8391779 0.9286542
ex3 <- flexmix(Nclus[, 1] ~ 1, cluster = cut(Nclus[, 1], 3),
model = FLXMCnorm1())
ex3
#>
#> Call:
#> flexmix(formula = Nclus[, 1] ~ 1, cluster = cut(Nclus[, 1], 3),
#> model = FLXMCnorm1())
#>
#> Cluster sizes:
#> 1 2 3
#> 262 186 102
#>
#> convergence after 128 iterations
parameters(ex3)
#> Comp.1 Comp.2 Comp.3
#> mean -0.9217948 4.0600622 7.9189585
#> sd 1.8563637 0.8391779 0.9286542