Random Number Generator for Finite Mixtures
rflexmix.RdGiven a finite mixture model generate random numbers from it.
rflexmix(object, newdata, ...)Arguments
- object
A fitted finite mixture model of class
flexmixor an unfitted of classFLXdist.- newdata
Optionally, a data frame in which to look for variables with which to predict or an integer specifying the number of random draws for model-based clustering. If omitted, the data to which the model was fitted is used.
- ...
Further arguments to be passed to or from methods.
Details
rflexmix provides the creation of the model matrix for new data
and the sampling of the cluster memberships. The sampling of the
component distributions given the classification is done by calling
rFLXM. This step has to be provided for the different model
classes.
Value
A list with components
- y
Random sample
- group
Grouping factor
- class
Class membership
Examples
example(flexmix)
#>
#> flexmx> data("NPreg", package = "flexmix")
#>
#> flexmx> ## mixture of two linear regression models. Note that control parameters
#> flexmx> ## can be specified as named list and abbreviated if unique.
#> flexmx> ex1 <- flexmix(yn ~ x + I(x^2), data = NPreg, k = 2,
#> flexmx+ control = list(verb = 5, iter = 100))
#> Classification: weighted
#> 5 Log-likelihood : -722.2404
#> 10 Log-likelihood : -656.0035
#> 15 Log-likelihood : -642.5457
#> 16 Log-likelihood : -642.5453
#> converged
#>
#> flexmx> ex1
#>
#> Call:
#> flexmix(formula = yn ~ x + I(x^2), data = NPreg, k = 2, control = list(verb = 5,
#> iter = 100))
#>
#> Cluster sizes:
#> 1 2
#> 100 100
#>
#> convergence after 16 iterations
#>
#> flexmx> summary(ex1)
#>
#> Call:
#> flexmix(formula = yn ~ x + I(x^2), data = NPreg, k = 2, control = list(verb = 5,
#> iter = 100))
#>
#> prior size post>0 ratio
#> Comp.1 0.494 100 145 0.690
#> Comp.2 0.506 100 141 0.709
#>
#> 'log Lik.' -642.5453 (df=9)
#> AIC: 1303.091 BIC: 1332.776
#>
#>
#> flexmx> plot(ex1)
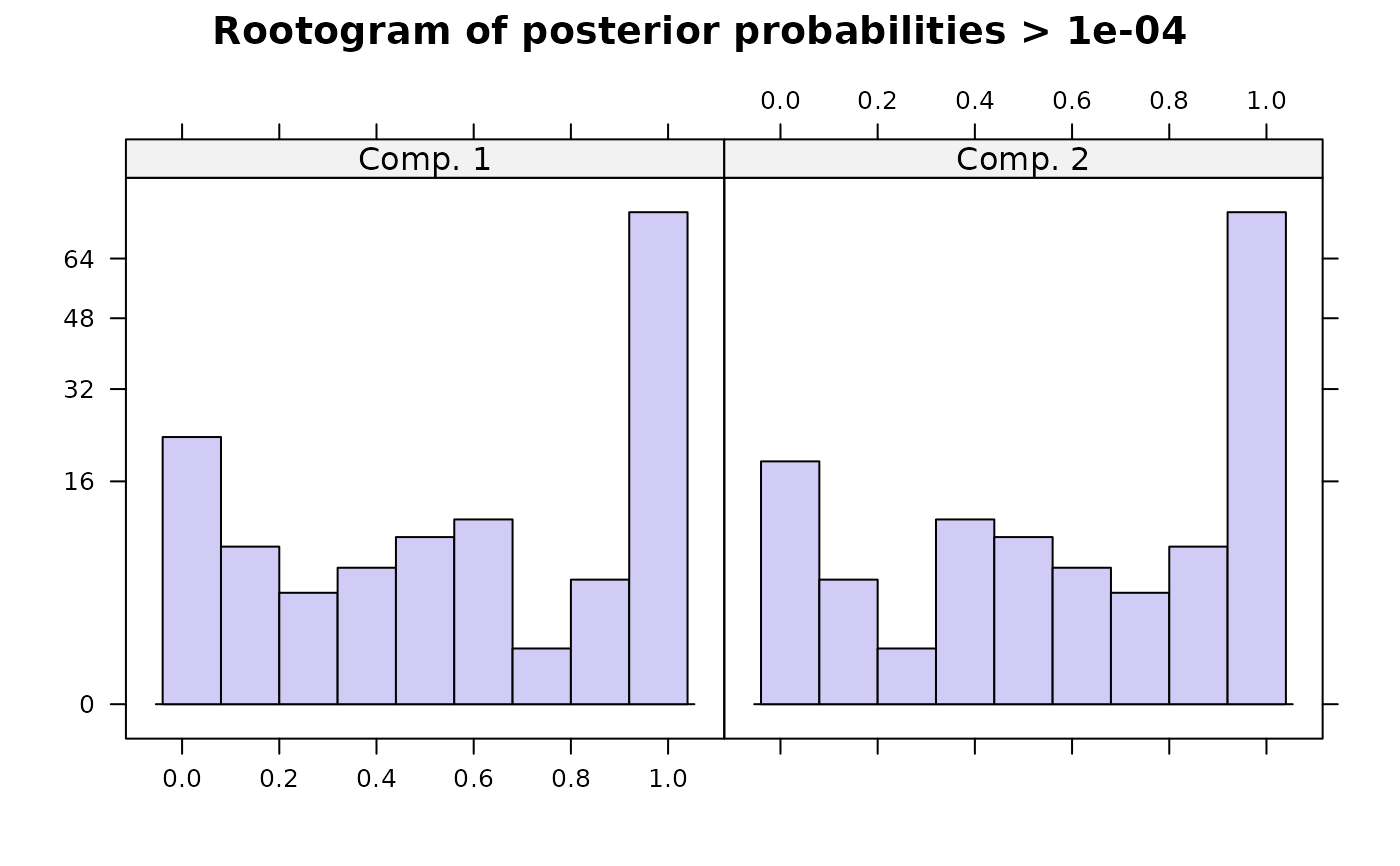 #>
#> flexmx> ## now we fit a model with one Gaussian response and one Poisson
#> flexmx> ## response. Note that the formulas inside the call to FLXMRglm are
#> flexmx> ## relative to the overall model formula.
#> flexmx> ex2 <- flexmix(yn ~ x, data = NPreg, k = 2,
#> flexmx+ model = list(FLXMRglm(yn ~ . + I(x^2)),
#> flexmx+ FLXMRglm(yp ~ ., family = "poisson")))
#>
#> flexmx> plot(ex2)
#>
#> flexmx> ## now we fit a model with one Gaussian response and one Poisson
#> flexmx> ## response. Note that the formulas inside the call to FLXMRglm are
#> flexmx> ## relative to the overall model formula.
#> flexmx> ex2 <- flexmix(yn ~ x, data = NPreg, k = 2,
#> flexmx+ model = list(FLXMRglm(yn ~ . + I(x^2)),
#> flexmx+ FLXMRglm(yp ~ ., family = "poisson")))
#>
#> flexmx> plot(ex2)
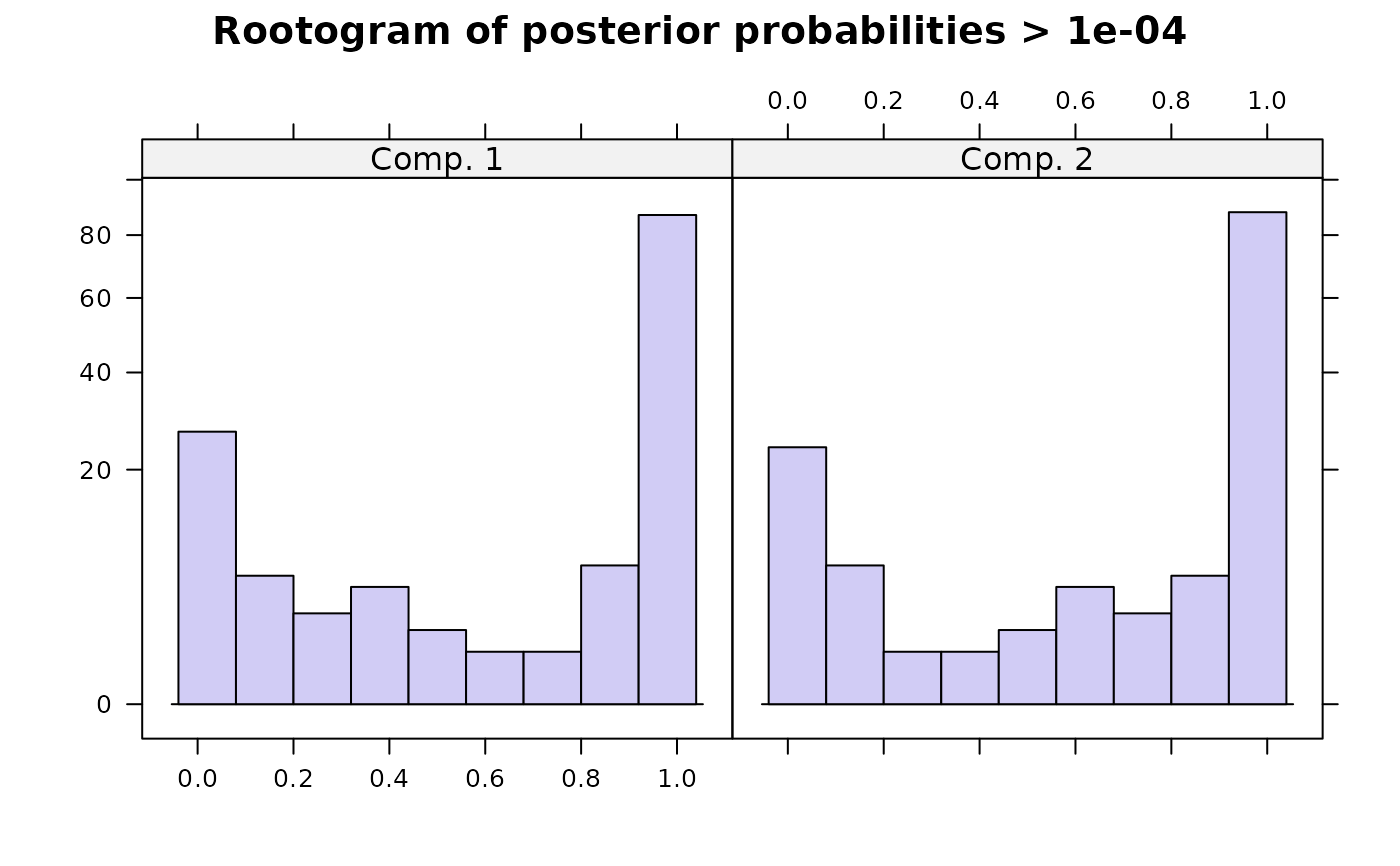 #>
#> flexmx> ex2
#>
#> Call:
#> flexmix(formula = yn ~ x, data = NPreg, k = 2, model = list(FLXMRglm(yn ~
#> . + I(x^2)), FLXMRglm(yp ~ ., family = "poisson")))
#>
#> Cluster sizes:
#> 1 2
#> 96 104
#>
#> convergence after 11 iterations
#>
#> flexmx> table(ex2@cluster, NPreg$class)
#>
#> 1 2
#> 1 1 95
#> 2 99 5
#>
#> flexmx> ## for Gaussian responses we get coefficients and standard deviation
#> flexmx> parameters(ex2, component = 1, model = 1)
#> Comp.1
#> coef.(Intercept) 14.5914766
#> coef.x 9.9146975
#> coef.I(x^2) -0.9756903
#> sigma 3.4115522
#>
#> flexmx> ## for Poisson response we get only coefficients
#> flexmx> parameters(ex2, component = 1, model = 2)
#> Comp.1
#> coef.(Intercept) 1.03770716
#> coef.x 0.09106366
#>
#> flexmx> ## fitting a model only to the Poisson response is of course
#> flexmx> ## done like this
#> flexmx> ex3 <- flexmix(yp ~ x, data = NPreg, k = 2,
#> flexmx+ model = FLXMRglm(family = "poisson"))
#>
#> flexmx> ## if observations are grouped, i.e., we have several observations per
#> flexmx> ## individual, fitting is usually much faster:
#> flexmx> ex4 <- flexmix(yp~x|id1, data = NPreg, k = 2,
#> flexmx+ model = FLXMRglm(family = "poisson"))
#>
#> flexmx> ## And now a binomial example. Mixtures of binomials are not generically
#> flexmx> ## identified, here the grouping variable is necessary:
#> flexmx> set.seed(1234)
#>
#> flexmx> ex5 <- initFlexmix(cbind(yb,1 - yb) ~ x, data = NPreg, k = 2,
#> flexmx+ model = FLXMRglm(family = "binomial"), nrep = 5)
#> 2 : * * * * *
#>
#> flexmx> table(NPreg$class, clusters(ex5))
#>
#> 1 2
#> 1 0 100
#> 2 9 91
#>
#> flexmx> ex6 <- initFlexmix(cbind(yb, 1 - yb) ~ x | id2, data = NPreg, k = 2,
#> flexmx+ model = FLXMRglm(family = "binomial"), nrep = 5)
#> 2 : * * * * *
#>
#> flexmx> table(NPreg$class, clusters(ex6))
#>
#> 1 2
#> 1 92 8
#> 2 4 96
sample <- rflexmix(ex1)
#>
#> flexmx> ex2
#>
#> Call:
#> flexmix(formula = yn ~ x, data = NPreg, k = 2, model = list(FLXMRglm(yn ~
#> . + I(x^2)), FLXMRglm(yp ~ ., family = "poisson")))
#>
#> Cluster sizes:
#> 1 2
#> 96 104
#>
#> convergence after 11 iterations
#>
#> flexmx> table(ex2@cluster, NPreg$class)
#>
#> 1 2
#> 1 1 95
#> 2 99 5
#>
#> flexmx> ## for Gaussian responses we get coefficients and standard deviation
#> flexmx> parameters(ex2, component = 1, model = 1)
#> Comp.1
#> coef.(Intercept) 14.5914766
#> coef.x 9.9146975
#> coef.I(x^2) -0.9756903
#> sigma 3.4115522
#>
#> flexmx> ## for Poisson response we get only coefficients
#> flexmx> parameters(ex2, component = 1, model = 2)
#> Comp.1
#> coef.(Intercept) 1.03770716
#> coef.x 0.09106366
#>
#> flexmx> ## fitting a model only to the Poisson response is of course
#> flexmx> ## done like this
#> flexmx> ex3 <- flexmix(yp ~ x, data = NPreg, k = 2,
#> flexmx+ model = FLXMRglm(family = "poisson"))
#>
#> flexmx> ## if observations are grouped, i.e., we have several observations per
#> flexmx> ## individual, fitting is usually much faster:
#> flexmx> ex4 <- flexmix(yp~x|id1, data = NPreg, k = 2,
#> flexmx+ model = FLXMRglm(family = "poisson"))
#>
#> flexmx> ## And now a binomial example. Mixtures of binomials are not generically
#> flexmx> ## identified, here the grouping variable is necessary:
#> flexmx> set.seed(1234)
#>
#> flexmx> ex5 <- initFlexmix(cbind(yb,1 - yb) ~ x, data = NPreg, k = 2,
#> flexmx+ model = FLXMRglm(family = "binomial"), nrep = 5)
#> 2 : * * * * *
#>
#> flexmx> table(NPreg$class, clusters(ex5))
#>
#> 1 2
#> 1 0 100
#> 2 9 91
#>
#> flexmx> ex6 <- initFlexmix(cbind(yb, 1 - yb) ~ x | id2, data = NPreg, k = 2,
#> flexmx+ model = FLXMRglm(family = "binomial"), nrep = 5)
#> 2 : * * * * *
#>
#> flexmx> table(NPreg$class, clusters(ex6))
#>
#> 1 2
#> 1 92 8
#> 2 4 96
sample <- rflexmix(ex1)