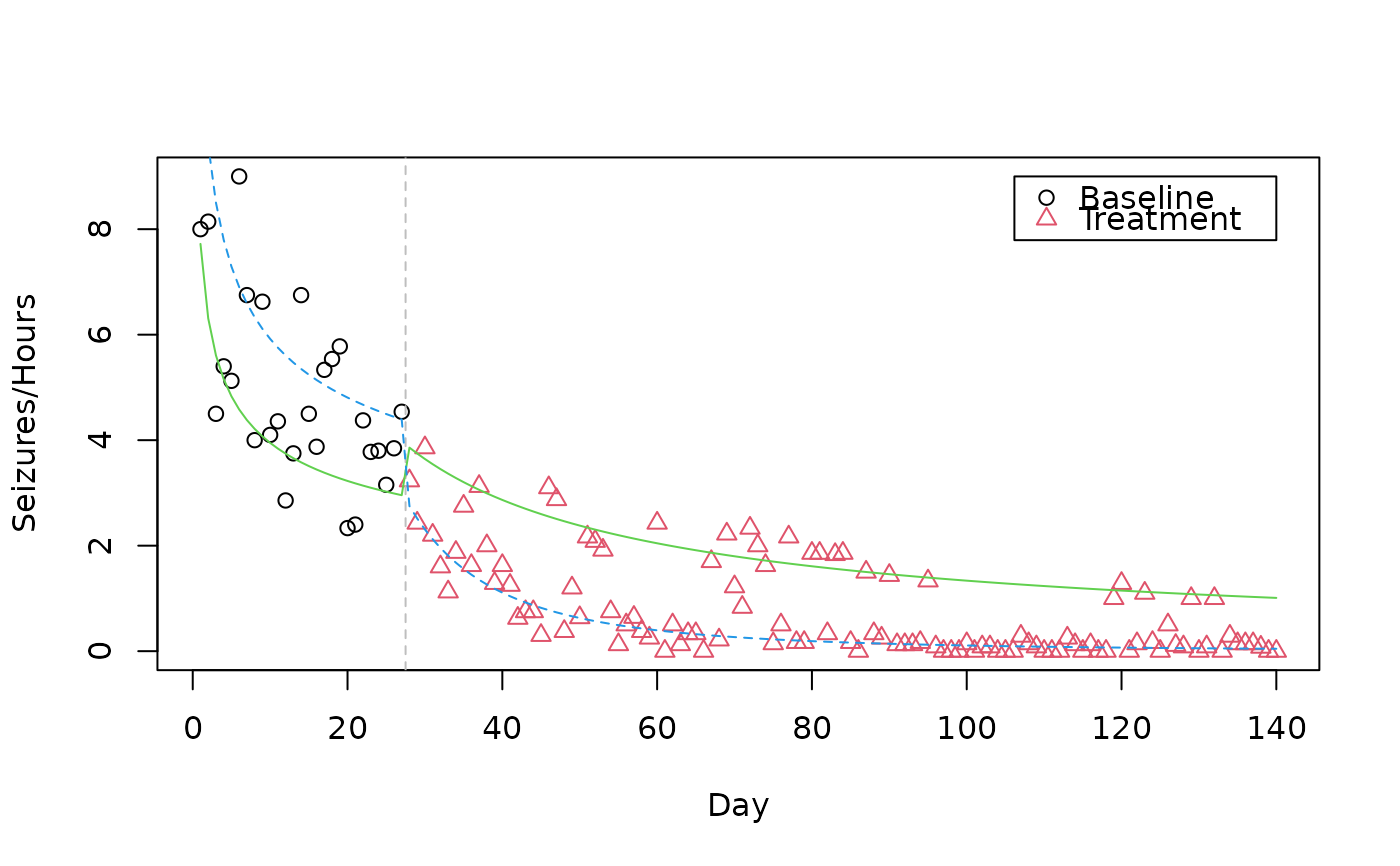
Epileptic Seizure Data
seizure.RdData from a clinical trial where the effect of intravenous gamma-globulin on suppression of epileptic seizures is studied. Daily observations for a period of 140 days on one patient are given, where the first 27 days are a baseline period without treatment, the remaining 113 days are the treatment period.
data("seizure")Format
A data frame with 140 observations on the following 4 variables.
- Seizures
A numeric vector, daily counts of epileptic seizures.
- Hours
A numeric vector, hours of daily parental observation.
- Treatment
A factor with levels
NoandYes.- Day
A numeric vector.
Source
P. Wang, M. Puterman, I. Cockburn, and N. Le. Mixed poisson regression models with covariate dependent rates. Biometrics, 52, 381–400, 1996.
References
B. Gruen and F. Leisch. Bootstrapping finite mixture models. In J. Antoch, editor, Compstat 2004–Proceedings in Computational Statistics, 1115–1122. Physika Verlag, Heidelberg, Germany, 2004. ISBN 3-7908-1554-3.
Examples
data("seizure", package = "flexmix")
plot(Seizures/Hours ~ Day, col = as.integer(Treatment),
pch = as.integer(Treatment), data = seizure)
abline(v = 27.5, lty = 2, col = "grey")
legend(140, 9, c("Baseline", "Treatment"),
pch = 1:2, col = 1:2, xjust = 1, yjust = 1)
set.seed(123)
## The model presented in the Wang et al paper: two components for
## "good" and "bad" days, respectively, each a Poisson GLM with hours of
## parental observation as offset
seizMix <- flexmix(Seizures ~ Treatment * log(Day),
data = seizure, k = 2,
model = FLXMRglm(family = "poisson",
offset = log(seizure$Hours)))
summary(seizMix)
#>
#> Call:
#> flexmix(formula = Seizures ~ Treatment * log(Day), data = seizure,
#> k = 2, model = FLXMRglm(family = "poisson", offset = log(seizure$Hours)))
#>
#> prior size post>0 ratio
#> Comp.1 0.307 42 108 0.389
#> Comp.2 0.693 98 116 0.845
#>
#> 'log Lik.' -380.7697 (df=9)
#> AIC: 779.5393 BIC: 806.0141
#>
summary(refit(seizMix))
#> $Comp.1
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 2.043989 0.120852 16.9132 < 2.2e-16 ***
#> TreatmentYes 2.081361 0.495066 4.2042 2.620e-05 ***
#> log(Day) -0.291203 0.057822 -5.0362 4.749e-07 ***
#> TreatmentYes:log(Day) -0.541596 0.128682 -4.2088 2.567e-05 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> $Comp.2
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 2.470428 0.242776 10.1757 < 2.2e-16 ***
#> TreatmentYes 7.025729 0.597897 11.7507 < 2.2e-16 ***
#> log(Day) -0.300391 0.086015 -3.4923 0.0004789 ***
#> TreatmentYes:log(Day) -2.245671 0.166680 -13.4729 < 2.2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
matplot(seizure$Day, fitted(seizMix)/seizure$Hours, type = "l",
add = TRUE, col = 3:4)