This function is used to separate and place individual
labels in their own rows if your variable names contain multiple
delimited labels.
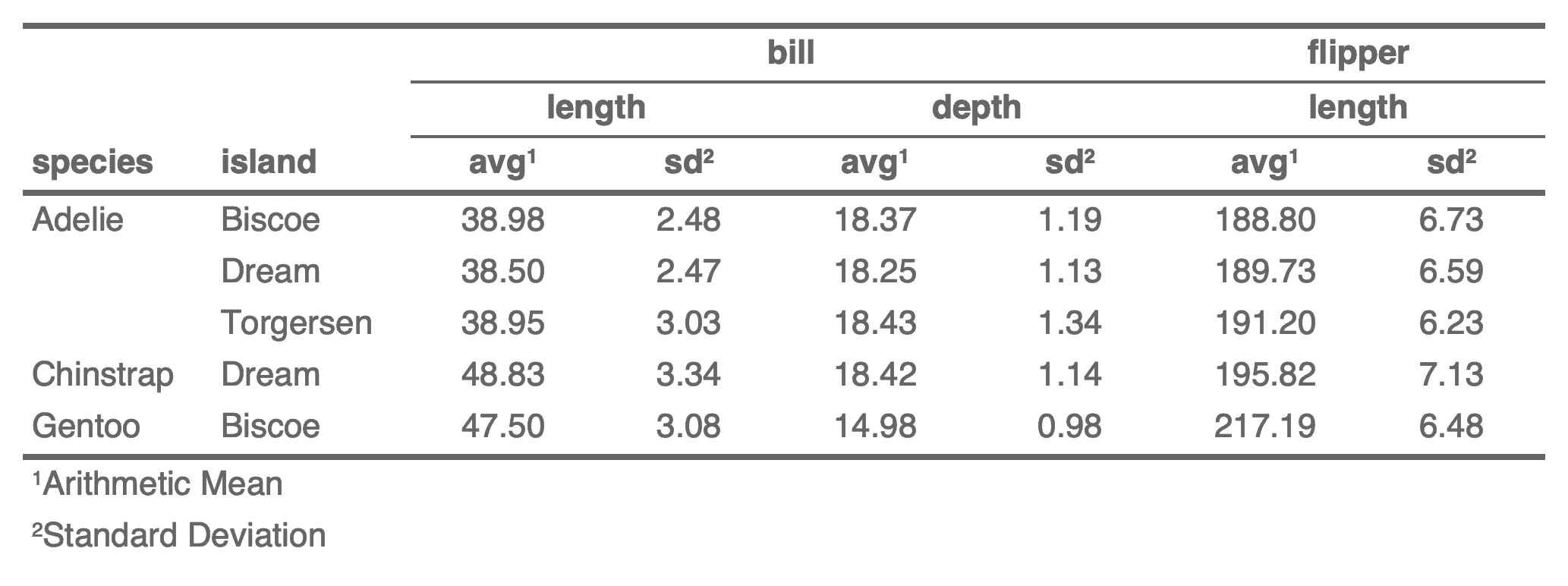
separate_header(
x,
opts = c("span-top", "center-hspan", "bottom-vspan", "default-theme"),
split = "[_\\.]",
fixed = FALSE
)Arguments
- x
a flextable object
- opts
Optional treatments to apply to the resulting header part. This should be a character vector with support for multiple values.
Supported values include:
"span-top": This operation spans empty cells with the first non-empty cell, applied column by column.
"center-hspan": Center the cells that are horizontally spanned.
"bottom-vspan": Aligns to the bottom the cells treated at the when "span-top" is applied.
"default-theme": Applies the theme set in
set_flextable_defaults(theme_fun = ...)to the new header part.
- split
a regular expression (unless
fixed = TRUE) to use for splitting.- fixed
logical. If TRUE match
splitexactly, otherwise use regular expressions.
See also
Other functions for row and column operations in a flextable:
add_body(),
add_body_row(),
add_footer(),
add_footer_lines(),
add_footer_row(),
add_header(),
add_header_row(),
delete_columns(),
delete_part(),
delete_rows(),
set_header_footer_df,
set_header_labels()
Examples
library(flextable)
x <- data.frame(
Species = as.factor(c("setosa", "versicolor", "virginica")),
Sepal.Length_mean = c(5.006, 5.936, 6.588),
Sepal.Length_sd = c(0.35249, 0.51617, 0.63588),
Sepal.Width_mean = c(3.428, 2.77, 2.974),
Sepal.Width_sd = c(0.37906, 0.3138, 0.3225),
Petal.Length_mean = c(1.462, 4.26, 5.552),
Petal.Length_sd = c(0.17366, 0.46991, 0.55189),
Petal.Width_mean = c(0.246, 1.326, 2.026),
Petal.Width_sd = c(0.10539, 0.19775, 0.27465)
)
ft_1 <- flextable(x)
ft_1 <- colformat_double(ft_1, digits = 2)
ft_1 <- theme_box(ft_1)
ft_1 <- separate_header(
x = ft_1,
opts = c("span-top", "bottom-vspan")
)
ft_1
Species
Sepal
Petal
Length
Width
Length
Width
mean
sd
mean
sd
mean
sd
mean
sd
setosa
5.01
0.35
3.43
0.38
1.46
0.17
0.25
0.11
versicolor
5.94
0.52
2.77
0.31
4.26
0.47
1.33
0.20
virginica
6.59
0.64
2.97
0.32
5.55
0.55
2.03
0.27