plot pedigree.shrink object that is a shrunk pedigree object
plot.pedigree.shrink.RdPlot the pedigree object that is the trimmed pedigree.shrink object along with colors based on availability and affection status.
# S3 method for class 'pedigree.shrink'
plot(x, bigped=FALSE, title="", xlegend="topright", ...)Arguments
- x
A pedigree.shrink object, which contains a pedigree object and information about which subject was removed.
- bigped
Logical value indicating whether pedigree should be compacted to fit in plotting region. If T, then packed=F is used in pedigree() along with smaller symbol sizes.
- title
Optional plot title
- xlegend
The x argument for the legend command, which allows coordinates or, more conveniently, options such as "topright", "right", "left", "bottomleft", etc., which is useful for pedigrees that cover most of the plot region.
- ...
Optional arguments to plot method
See also
Examples
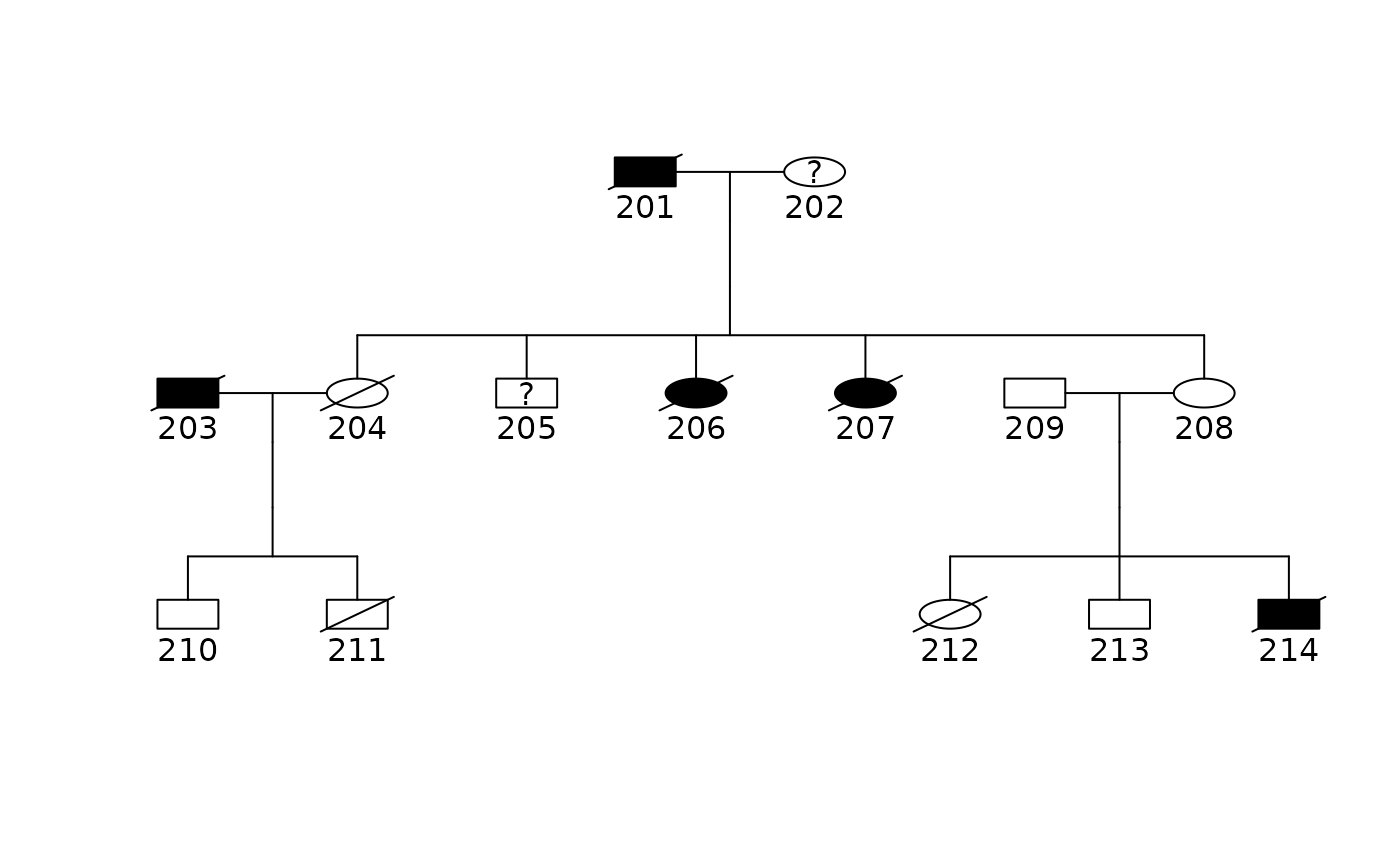
data(sample.ped)
fam2 <- sample.ped[sample.ped$ped==2,]
ped2 <- pedigree(fam2$id, fam2$father, fam2$mother, fam2$sex,
fam2$affected, fam2$avail)
shrink2 <- pedigree.shrink(ped2,avail=fam2$avail)
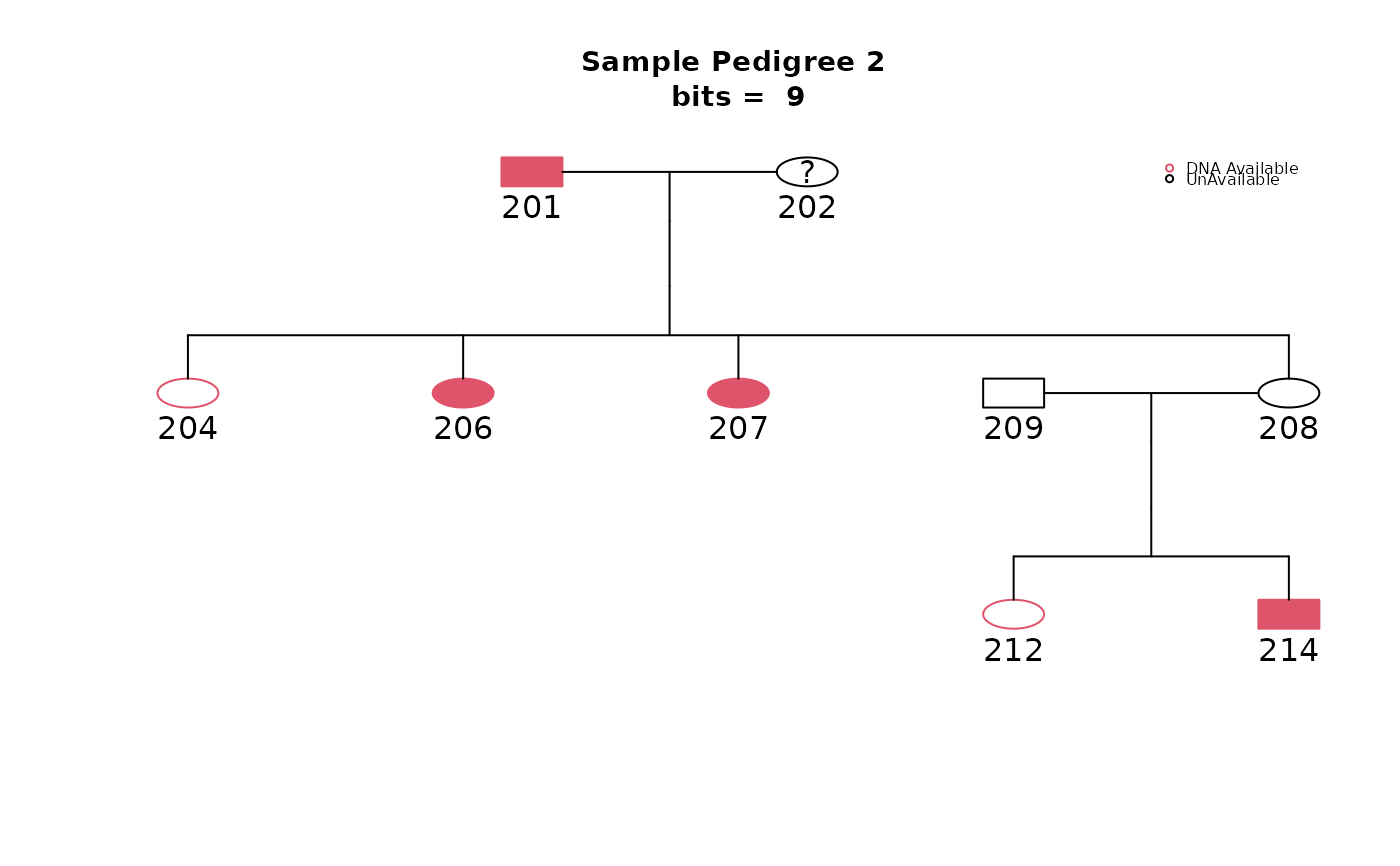
shrink2
#> Pedigree Size:
#> N.subj Bits
#> Original 14 16
#> Only Informative 9 9
#> Trimmed 9 9
#>
#> Unavailable subjects trimmed:
#> 205 210 213
#>
#> Non-informative subjects trimmed:
#> 203 211
plot(ped2)
 plot.pedigree.shrink(shrink2, title="Sample Pedigree 2")
plot.pedigree.shrink(shrink2, title="Sample Pedigree 2")