Trellis Displays of Tukey's Hanging Rootograms
rootogram.RdDisplays hanging rootograms.
rootogram(x, ...)
# S3 method for class 'formula'
rootogram(x, data = parent.frame(),
ylab = expression(sqrt(P(X == x))),
prepanel = prepanel.rootogram,
panel = panel.rootogram,
...,
probability = TRUE)
prepanel.rootogram(x, y = table(x),
dfun = NULL,
transformation = sqrt,
hang = TRUE,
probability = TRUE,
...)
panel.rootogram(x, y = table(x),
dfun = NULL,
col = plot.line$col,
lty = plot.line$lty,
lwd = plot.line$lwd,
alpha = plot.line$alpha,
transformation = sqrt,
hang = TRUE,
probability = TRUE,
type = "l", pch = 16,
...)Arguments
- x, y
For
rootogram,xis the object on which method dispatch is carried out. For the"formula"method,xis a formula describing the form of conditioning plot. The formula can be either of the form~xor of the formy~x. In the first case,xis assumed to be a vector of raw observations, and an observed frequency distribution is computed from it. In the second case,xis assumed to be unique values andythe corresponding frequencies. In either case, further conditioning variables are allowed.A similar interpretation holds for
xandyinprepanel.rootogramandpanel.rootogram.Note that the data are assumed to arise from a discrete distribution with some probability mass function. See details below.
- data
For the
"formula"method, a data frame containing values for any variables in the formula, as well as those ingroupsandsubsetif applicable (groupsis currently ignored by the default panel function). By default the environment where the function was called from is used.- dfun
a probability mass function, to be evaluated at unique x values
- prepanel, panel
panel and prepanel function used to create the display.
- ylab
the y-axis label; typically a character string or an expression.
- col, lty, lwd, alpha
graphical parameters
- transformation
a vectorized function. Relative frequencies (observed) and theoretical probabilities (
dfun) are transformed by this function before being plotted.- hang
logical, whether lines representing observed relative freuqncies should “hang” from the curve representing the theoretical probabilities.
- probability
A logical flag, controlling whether the y-values are to be standardized to be probabilities by dividing by their sum.
- type
A character vector consisting of one or both of
"p"and"l". If"p"is included, the evaluated values ofdfunwill be denoted by points, and if"l"is included, they will be joined by lines.- pch
The plotting character to be used for the
"p"type.- ...
extra arguments, passed on as appropriate. Standard lattice arguments as well as arguments to
panel.rootogramcan be supplied directly in the high levelrootogramcall.
Details
This function implements Tukey's hanging rootograms. As implemented,
rootogram assumes that the data arise from a discrete
distribution (either supplied in raw form, when y is
unspecified, or in terms of the frequency distribution) with some
unknown probability mass function (p.m.f.). The purpose of the plot
is to check whether the supplied theoretical p.m.f. dfun is a
reasonable fit for the data.
It is reasonable to consider rootograms for continuous data by
discretizing it (similar to a histogram), but this must be done by the
user before calling rootogram. An example is given below.
Also consider the rootogram function in the vcd package,
especially if the number of unique values is small.
Value
rootogram produces an object of class "trellis". The
update method can be used to update components of the object and
the print method (usually called by default) will plot it on an
appropriate plotting device.
References
John W. Tukey (1972) Some graphic and semi-graphic displays. In T. A. Bancroft (Ed) Statistical Papers in Honor of George W. Snedecor, pp. 293–316. Available online at https://www.edwardtufte.com/tufte/tukey
See also
Examples
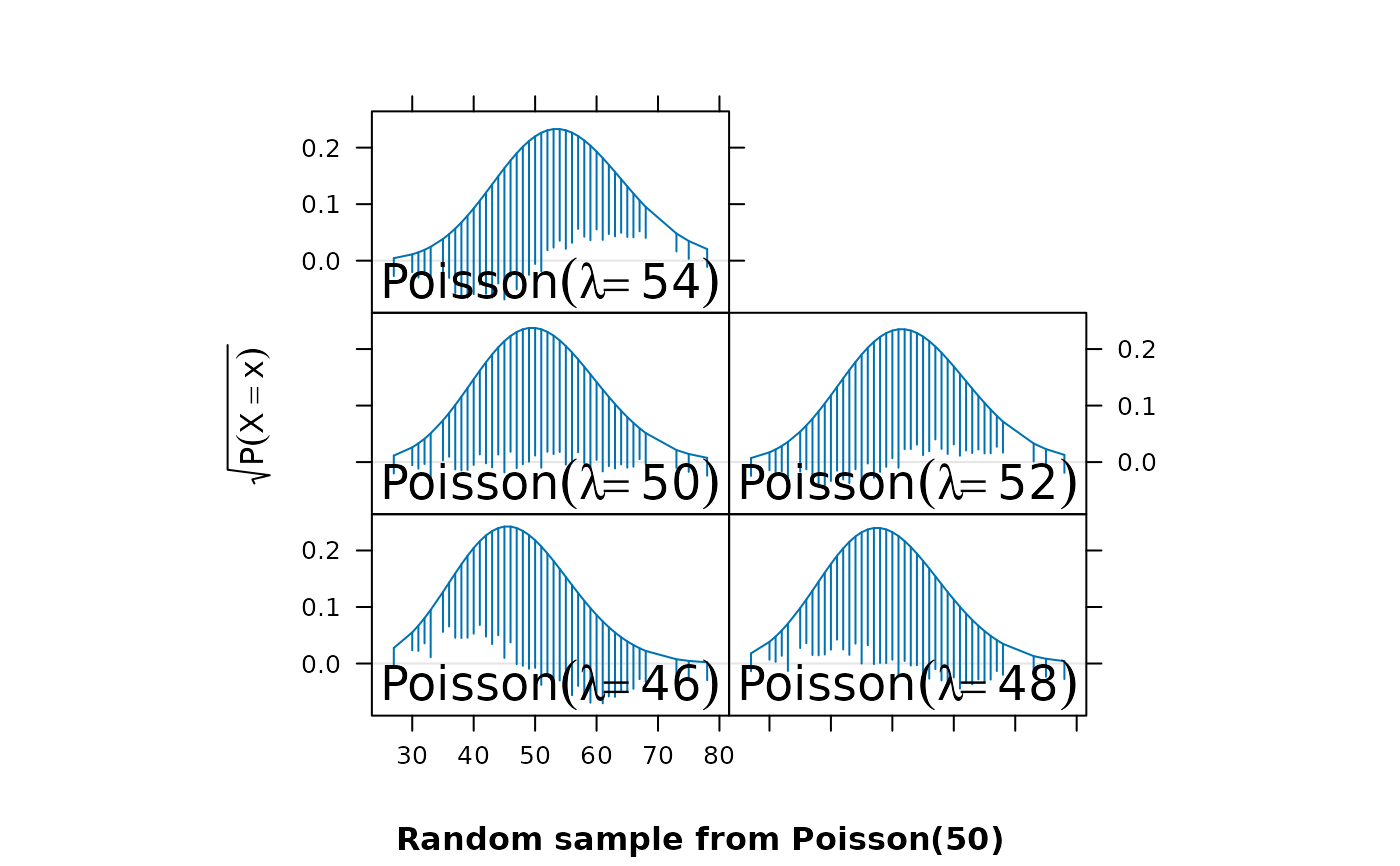
library(lattice)
x <- rpois(1000, lambda = 50)
p <- rootogram(~x, dfun = function(x) dpois(x, lambda = 50))
p
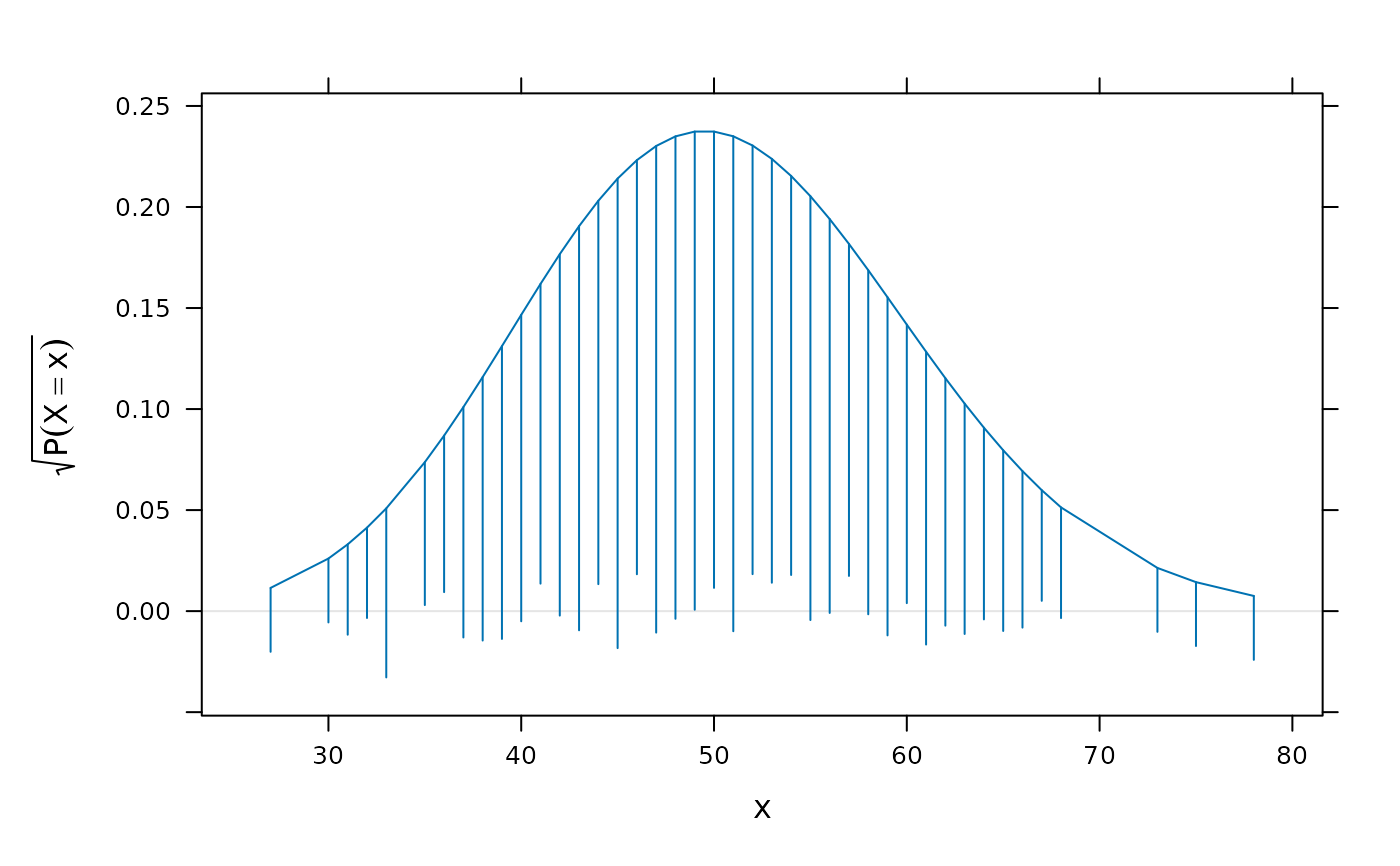 lambdav <- c(30, 40, 50, 60, 70)
update(p[rep(1, length(lambdav))],
aspect = "xy",
panel = function(x, ...) {
panel.rootogram(x,
dfun = function(x)
dpois(x, lambda = lambdav[panel.number()]))
})
lambdav <- c(30, 40, 50, 60, 70)
update(p[rep(1, length(lambdav))],
aspect = "xy",
panel = function(x, ...) {
panel.rootogram(x,
dfun = function(x)
dpois(x, lambda = lambdav[panel.number()]))
})
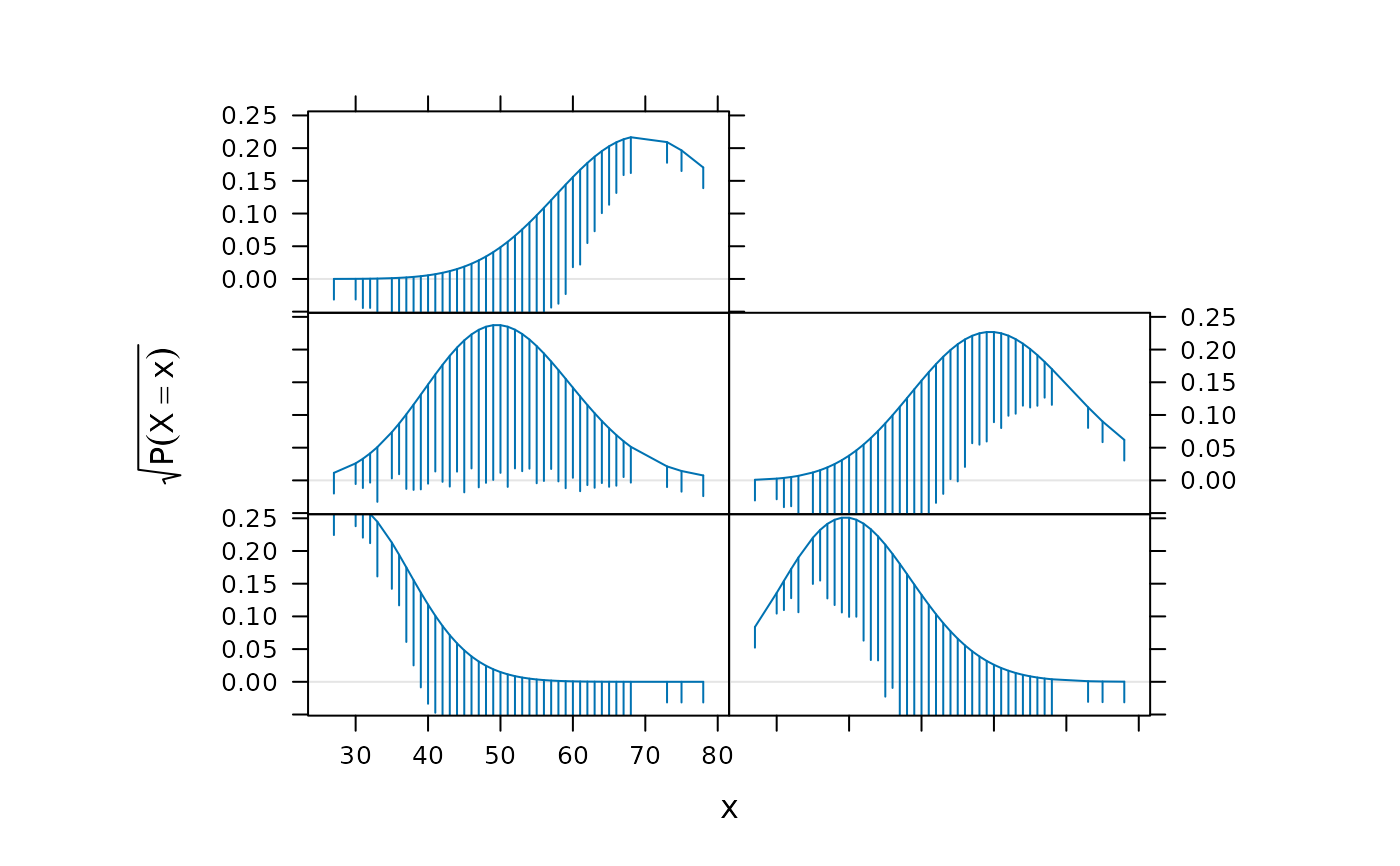 lambdav <- c(46, 48, 50, 52, 54)
update(p[rep(1, length(lambdav))],
aspect = "xy",
prepanel = function(x, ...) {
tmp <-
lapply(lambdav,
function(lambda) {
prepanel.rootogram(x,
dfun = function(x)
dpois(x, lambda = lambda))
})
list(xlim = range(sapply(tmp, "[[", "xlim")),
ylim = range(sapply(tmp, "[[", "ylim")),
dx = do.call("c", lapply(tmp, "[[", "dx")),
dy = do.call("c", lapply(tmp, "[[", "dy")))
},
panel = function(x, ...) {
panel.rootogram(x,
dfun = function(x)
dpois(x, lambda = lambdav[panel.number()]))
grid::grid.text(bquote(Poisson(lambda == .(foo)),
where = list(foo = lambdav[panel.number()])),
y = 0.15,
gp = grid::gpar(cex = 1.5))
},
xlab = "",
sub = "Random sample from Poisson(50)")
lambdav <- c(46, 48, 50, 52, 54)
update(p[rep(1, length(lambdav))],
aspect = "xy",
prepanel = function(x, ...) {
tmp <-
lapply(lambdav,
function(lambda) {
prepanel.rootogram(x,
dfun = function(x)
dpois(x, lambda = lambda))
})
list(xlim = range(sapply(tmp, "[[", "xlim")),
ylim = range(sapply(tmp, "[[", "ylim")),
dx = do.call("c", lapply(tmp, "[[", "dx")),
dy = do.call("c", lapply(tmp, "[[", "dy")))
},
panel = function(x, ...) {
panel.rootogram(x,
dfun = function(x)
dpois(x, lambda = lambdav[panel.number()]))
grid::grid.text(bquote(Poisson(lambda == .(foo)),
where = list(foo = lambdav[panel.number()])),
y = 0.15,
gp = grid::gpar(cex = 1.5))
},
xlab = "",
sub = "Random sample from Poisson(50)")
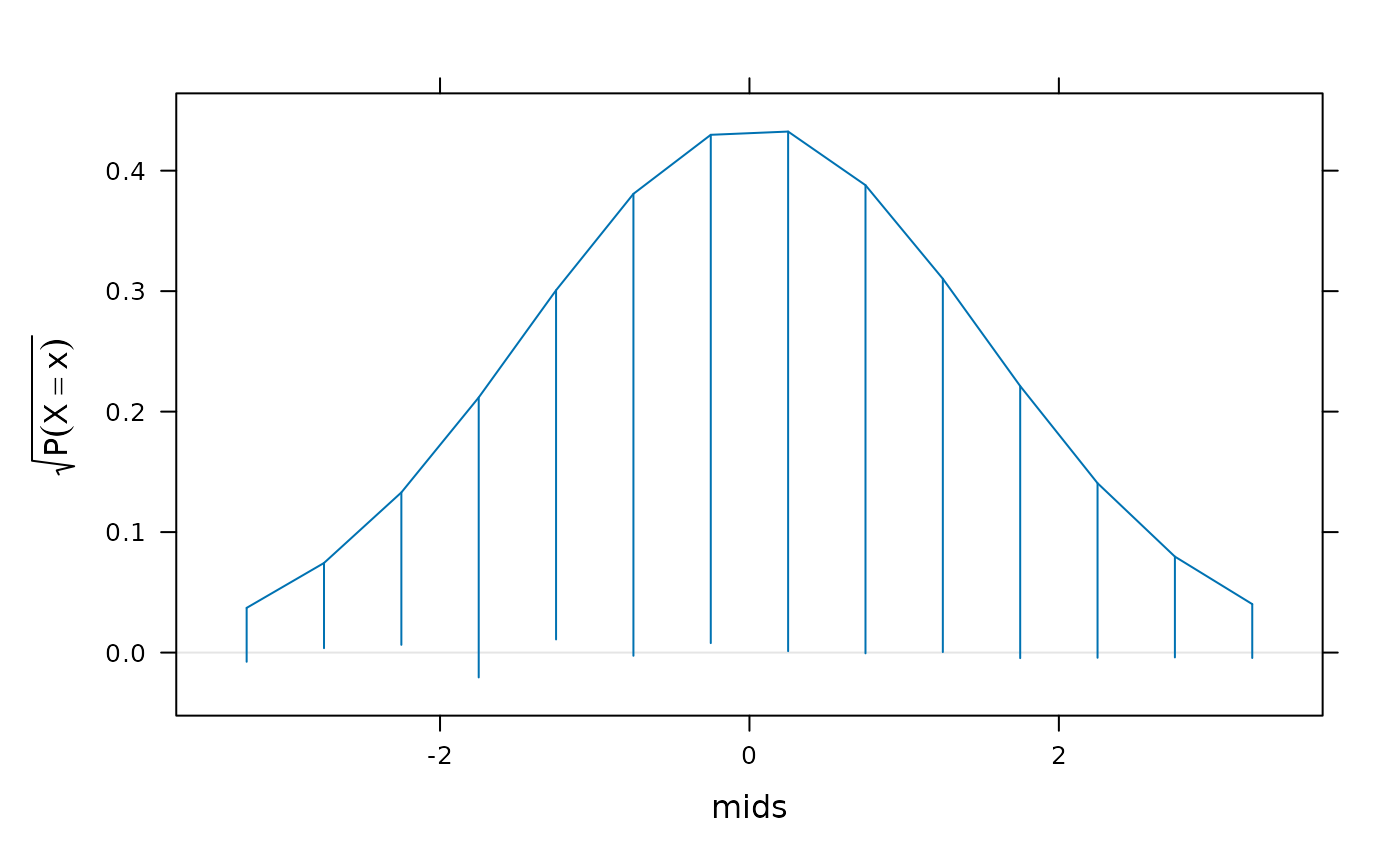
 ## Example using continuous data
xnorm <- rnorm(1000)
## 'discretize' by binning and replacing data by bin midpoints
h <- hist(xnorm, plot = FALSE)
## Option 1: Assume bin probabilities proportional to dnorm()
norm.factor <- sum(dnorm(h$mids, mean(xnorm), sd(xnorm)))
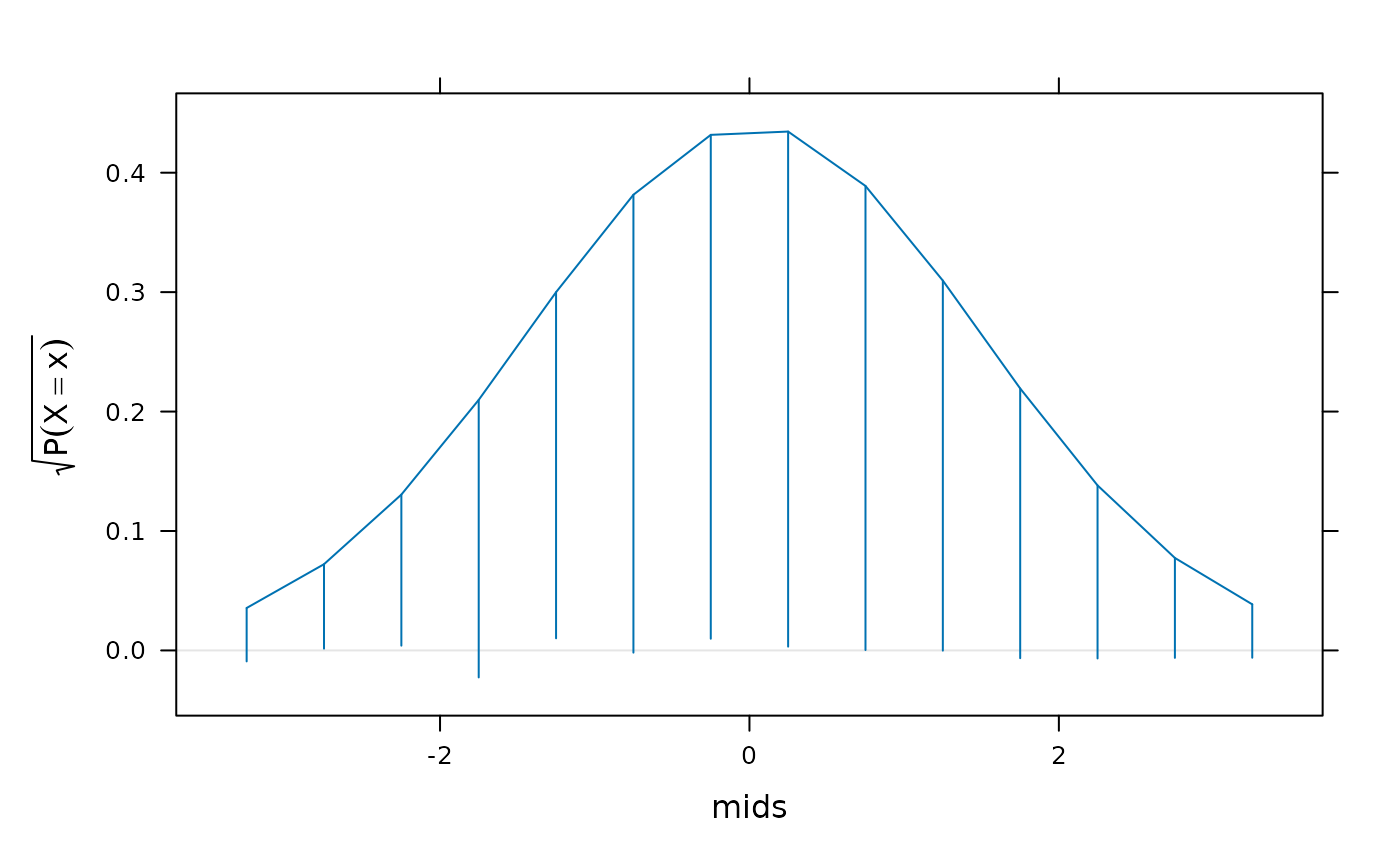
rootogram(counts ~ mids, data = h,
dfun = function(x) {
dnorm(x, mean(xnorm), sd(xnorm)) / norm.factor
})
## Example using continuous data
xnorm <- rnorm(1000)
## 'discretize' by binning and replacing data by bin midpoints
h <- hist(xnorm, plot = FALSE)
## Option 1: Assume bin probabilities proportional to dnorm()
norm.factor <- sum(dnorm(h$mids, mean(xnorm), sd(xnorm)))
rootogram(counts ~ mids, data = h,
dfun = function(x) {
dnorm(x, mean(xnorm), sd(xnorm)) / norm.factor
})
 ## Option 2: Compute probabilities explicitly using pnorm()
pdisc <- diff(pnorm(h$breaks, mean = mean(xnorm), sd = sd(xnorm)))
pdisc <- pdisc / sum(pdisc)
rootogram(counts ~ mids, data = h,
dfun = function(x) {
f <- factor(x, levels = h$mids)
pdisc[f]
})
## Option 2: Compute probabilities explicitly using pnorm()
pdisc <- diff(pnorm(h$breaks, mean = mean(xnorm), sd = sd(xnorm)))
pdisc <- pdisc / sum(pdisc)
rootogram(counts ~ mids, data = h,
dfun = function(x) {
f <- factor(x, levels = h$mids)
pdisc[f]
})