Hazard Ratio Plot
hazard.ratio.plot.RdThe hazard.ratio.plot function repeatedly estimates Cox
regression coefficients and confidence limits within time intervals.
The log hazard ratios are plotted against the mean failure/censoring
time within the interval. Unless times is specified, the number of
time intervals will be \(\max(round(d/e),2)\), where \(d\) is the
total number
of events in the sample. Efron's likelihood is used for estimating
Cox regression coefficients (using coxph.fit). In the case of
tied failure times, some intervals may have a point in common.
hazard.ratio.plot(x, Srv, which, times=, e=30, subset,
conf.int=.95, legendloc=NULL, smooth=TRUE, pr=FALSE, pl=TRUE,
add=FALSE, ylim, cex=.5, xlab="t", ylab, antilog=FALSE, ...)Arguments
- x
a vector or matrix of predictors
- Srv
a
Survobject- which
a vector of column numbers of
xfor which to estimate hazard ratios across time and make plots. The default is to do so for all predictors. Whenever one predictor is displayed, all other predictors in thexmatrix are adjusted for (with a separate adjustment form for each time interval).- times
optional vector of time interval endpoints. Example:
times=c(1,2,3)uses intervals[0,1), [1,2), [2,3), [3+). If times is omitted, uses intervals containingeevents- e
number of events per time interval if times not given
- subset
vector used for subsetting the entire analysis, e.g.
subset=sex=="female"- conf.int
confidence interval coverage
- legendloc
location for legend. Omit to use mouse,
"none"for none,"ll"for lower left of graph, or actual x and y coordinates (e.g.c(2,3))- smooth
also plot the super–smoothed version of the log hazard ratios
- pr
defaults to
FALSEto suppress printing of individual Cox fits- pl
defaults to
TRUEto plot results- add
add this plot to an already existing plot
- ylim
vector of
y-axis limits. Default is computed to include confidence bands.- cex
character size for legend information, default is 0.5
- xlab
label for
x-axis, default is"t"- ylab
label for
y-axis, default is"Log Hazard Ratio"or"Hazard Ratio", depending onantilog.- antilog
default is
FALSE. Set toTRUEto plot anti-log, i.e., hazard ratio.- ...
optional graphical parameters
See also
Examples
require(survival)
n <- 500
set.seed(1)
age <- 50 + 12*rnorm(n)
cens <- 15*runif(n)
h <- .02*exp(.04*(age-50))
d.time <- -log(runif(n))/h
label(d.time) <- 'Follow-up Time'
e <- ifelse(d.time <= cens,1,0)
d.time <- pmin(d.time, cens)
units(d.time) <- "Year"
hazard.ratio.plot(age, Surv(d.time,e), e=20, legendloc='ll')
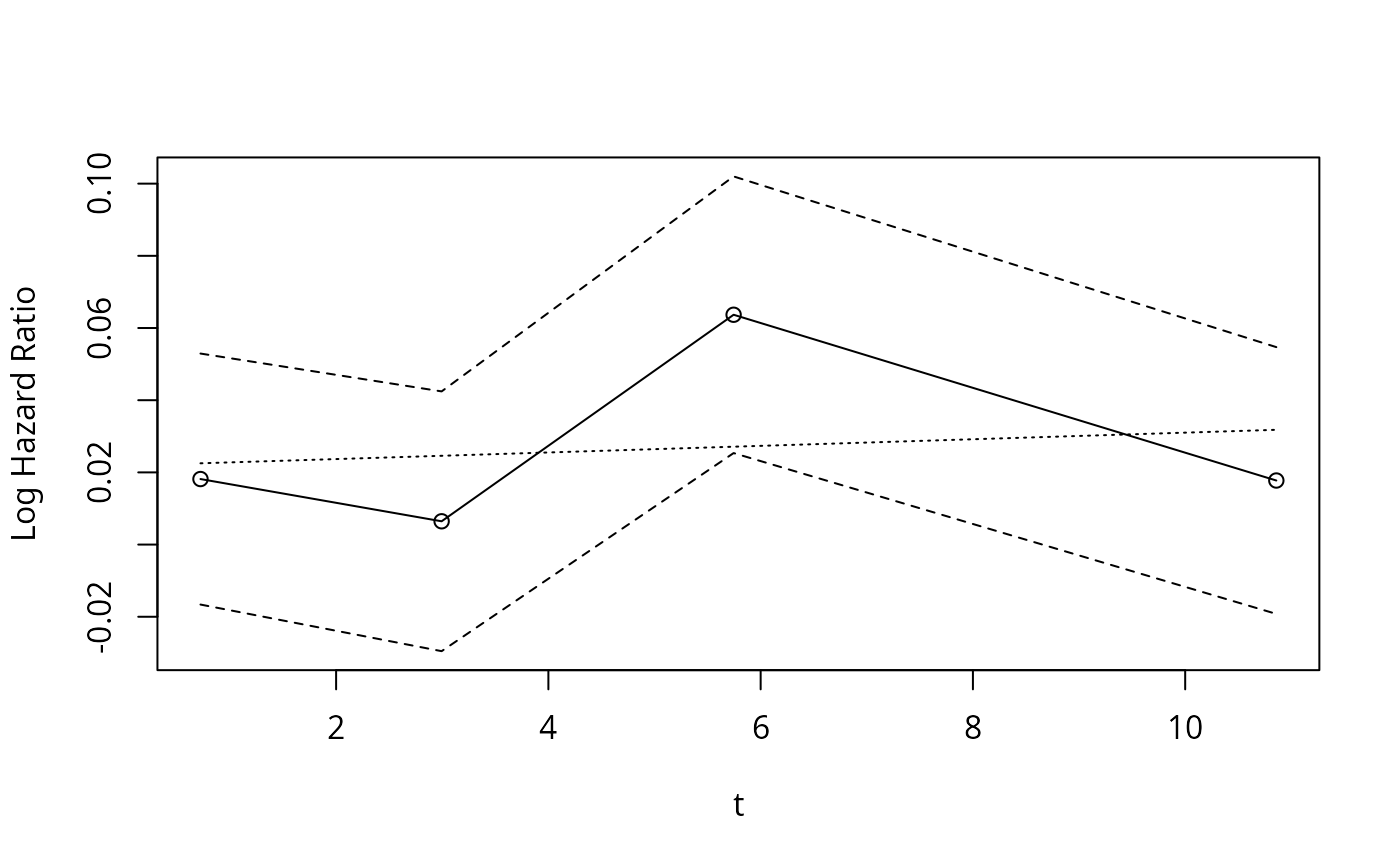 #> $time
#> [1] 0.72199 2.99413 5.74525 10.85871
#>
#> $log.hazard.ratio
#> [,1] [,2] [,3] [,4]
#> x1 0.018155 0.0064516 0.063675 0.017749
#>
#> $se
#> [,1] [,2] [,3] [,4]
#> [1,] 0.01774 0.01836 0.019553 0.018855
#>
#> $time
#> [1] 0.72199 2.99413 5.74525 10.85871
#>
#> $log.hazard.ratio
#> [,1] [,2] [,3] [,4]
#> x1 0.018155 0.0064516 0.063675 0.017749
#>
#> $se
#> [,1] [,2] [,3] [,4]
#> [1,] 0.01774 0.01836 0.019553 0.018855
#>