Plot method for model parameters
Source:R/plot.parameters_model.R, R/plot.parameters_sem.R
plot.see_parameters_model.RdThe plot() method for the parameters::model_parameters() function.
# S3 method for class 'see_parameters_model'
plot(
x,
show_intercept = FALSE,
size_point = 0.8,
size_text = NA,
sort = NULL,
n_columns = NULL,
type = c("forest", "funnel"),
weight_points = TRUE,
show_labels = FALSE,
show_estimate = TRUE,
show_interval = TRUE,
show_density = FALSE,
show_direction = TRUE,
log_scale = FALSE,
...
)
# S3 method for class 'see_parameters_sem'
plot(
x,
data = NULL,
component = c("regression", "correlation", "loading"),
type = component,
threshold_coefficient = NULL,
threshold_p = NULL,
ci = TRUE,
size_point = 22,
...
)Arguments
- x
An object.
- show_intercept
Logical, if
TRUE, the intercept-parameter is included in the plot. By default, it is hidden because in many cases the intercept-parameter has a posterior distribution on a very different location, so density curves of posterior distributions for other parameters are hardly visible.- size_point
Numeric specifying size of point-geoms.
- size_text
Numeric value specifying size of text labels.
- sort
The behavior of this argument depends on the plotting contexts.
Plotting model parameters: If
NULL, coefficients are plotted in the order as they appear in the summary. Settingsort = "ascending"orsort = "descending"sorts coefficients in ascending or descending order, respectively. Settingsort = TRUEis the same assort = "ascending".Plotting Bayes factors: Sort pie-slices by posterior probability (descending)?
- n_columns
For models with multiple components (like fixed and random, count and zero-inflated), defines the number of columns for the panel-layout. If
NULL, a single, integrated plot is shown.- type
Character indicating the type of plot. Only applies for model parameters from meta-analysis objects (e.g. metafor).
- weight_points
Logical. If
TRUE, for meta-analysis objects, point size will be adjusted according to the study-weights.- show_labels
Logical. If
TRUE, text labels are displayed.- show_estimate
Should the point estimate of each parameter be shown? (default:
TRUE)- show_interval
Should the compatibility interval(s) of each parameter be shown? (default:
TRUE)- show_density
Should the compatibility density (i.e., posterior, bootstrap, or confidence density) of each parameter be shown? (default:
FALSE)- show_direction
Should the "direction" of coefficients (e.g., positive or negative coefficients) be highlighted using different colors? (default:
TRUE)- log_scale
Should exponentiated coefficients (e.g., odds-ratios) be plotted on a log scale? (default:
FALSE)- ...
Arguments passed to or from other methods.
- data
The original data used to create this object. Can be a statistical model.
- component
Character indicating which component of the model should be plotted.
- threshold_coefficient
Numeric, threshold at which value coefficients will be displayed.
- threshold_p
Numeric, threshold at which value p-values will be displayed.
- ci
Logical, whether confidence intervals should be added to the plot.
Value
A ggplot2-object.
Note
By default, coefficients and their confidence intervals are colored
depending on whether they show a "positive" or "negative" association with
the outcome. E.g., in case of linear models, colors simply distinguish positive
or negative coefficients. For logistic regression models that are shown on the
odds ratio scale, colors distinguish odds ratios above or below 1. Use
show_direction = FALSE to disable this feature and only show a one-colored
forest plot.
Examples
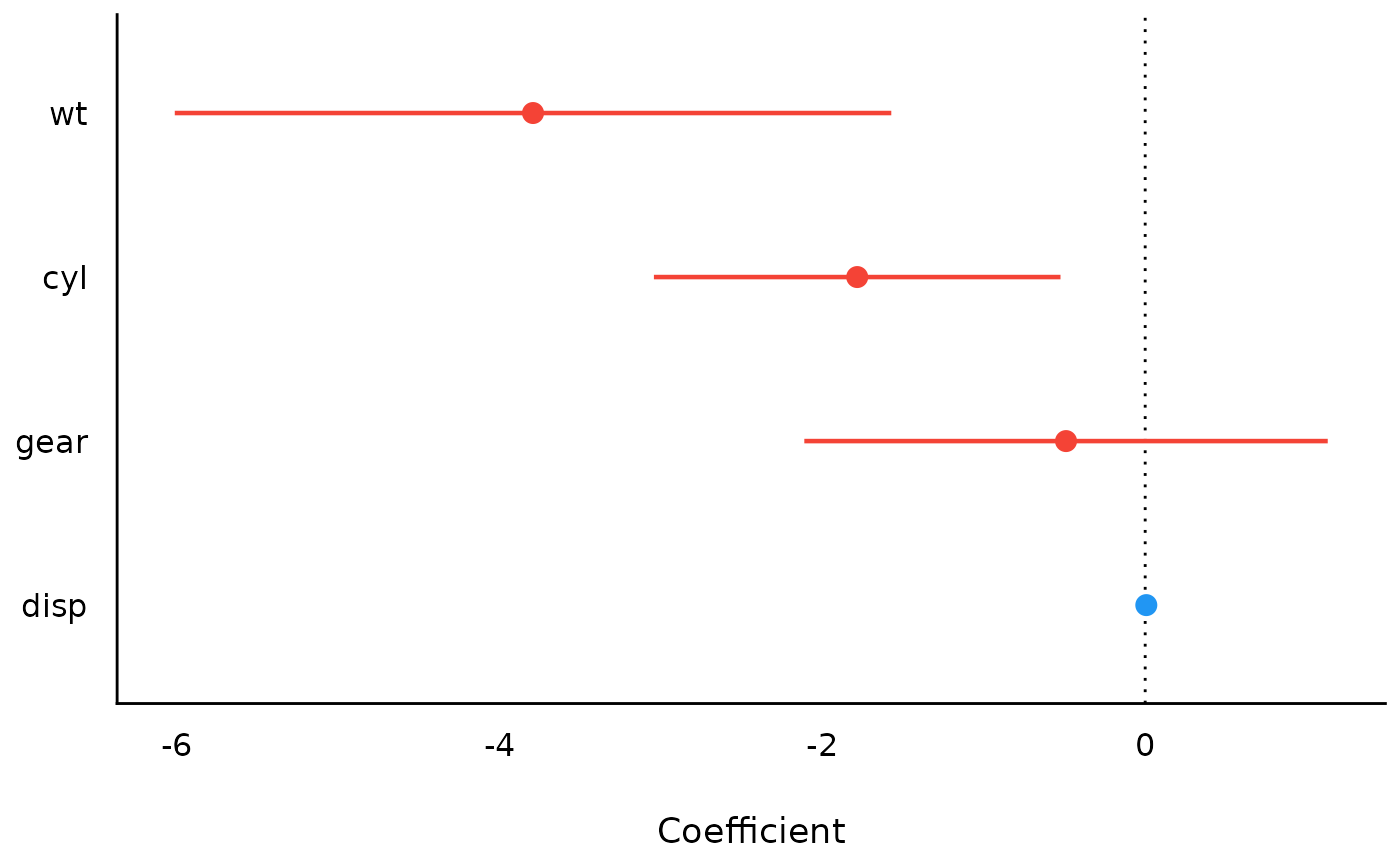
library(parameters)
m <- lm(mpg ~ wt + cyl + gear + disp, data = mtcars)
result <- model_parameters(m)
result
#> Parameter | Coefficient | SE | 95% CI | t(27) | p
#> ------------------------------------------------------------------
#> (Intercept) | 43.54 | 4.86 | [33.57, 53.51] | 8.96 | < .001
#> wt | -3.79 | 1.08 | [-6.01, -1.57] | -3.51 | 0.002
#> cyl | -1.78 | 0.61 | [-3.04, -0.52] | -2.91 | 0.007
#> gear | -0.49 | 0.79 | [-2.11, 1.13] | -0.62 | 0.540
#> disp | 6.94e-03 | 0.01 | [-0.02, 0.03] | 0.58 | 0.568
#>
#> Uncertainty intervals (equal-tailed) and p-values (two-tailed) computed
#> using a Wald t-distribution approximation.
plot(result)