Sequence Covering the Range of X, including X
seqXtend.RdProduce a sequence of unique values (sorted increasingly),
containing the initial set of values x.
This can be useful for setting prediction e.g. ranges in nonparametric
regression.
seqXtend(x, length., method = c("simple", "aim", "interpolate"),
from = NULL, to = NULL)Arguments
- x
numeric vector.
- length.
integer specifying approximately the desired
length()of the result.- method
string specifying the method to be used. The default,
"simple"usesseq(*, length.out = length.)where"aim"aims a bit better towards the desired final length, and"interpolate"interpolates evenly inside each interval \([x_i, x_{i+1}]\) in a way to make all the new intervalls of approximately the same length.- from, to
numbers to be passed to (the default method for)
seq(), defaulting to the minimal and maximalxvalue, respectively.
Note
method = "interpolate" typically gives the best results. Calling
roundfixS, it also need more computational resources
than the other methods.
Value
numeric vector of increasing values, of approximate length
length.
(unless length. < length(unique(x)) in which case, the result
is simply sort(unique(x))),
containing the original values of x.
From, r <- seqXtend(x, *), the original values are at
indices ix <- match(x,r), i.e., identical(x, r[ix]).
Examples
a <- c(1,2,10,12)
seqXtend(a, 12)# --> simply 1:12
#> [1] 1 2 3 4 5 6 7 8 9 10 11 12
seqXtend(a, 12, "interp")# ditto
#> [1] 1 2 3 4 5 6 7 8 9 10 11 12
seqXtend(a, 12, "aim")# really worse
#> [1] 1.00 2.00 2.22 3.44 4.67 5.89 7.11 8.33 9.56 10.00 10.78 12.00
stopifnot(all.equal(seqXtend(a, 12, "interp"), 1:12))
## for a "general" x, however, "aim" aims better than default
x <- c(1.2, 2.4, 4.6, 9.9)
length(print(seqXtend(x, 12))) # 14
#> [1] 1.20 1.99 2.40 2.78 3.57 4.36 4.60 5.15 5.95 6.74 7.53 8.32 9.11 9.90
#> [1] 14
length(print(seqXtend(x, 12, "aim"))) # 12
#> [1] 1.20 2.17 2.40 3.13 4.10 4.60 5.07 6.03 7.00 7.97 8.93 9.90
#> [1] 12
length(print(seqXtend(x, 12, "int"))) # 12
#> [1] 1.20 2.40 3.13 3.87 4.60 5.36 6.11 6.87 7.63 8.39 9.14 9.90
#> [1] 12
## "interpolate" is really nice:
xt <- seqXtend(x, 100, "interp")
plot(xt, main="seqXtend(*, 100, \"interpol\")")
points(match(x,xt), x, col = 2, pch = 20)
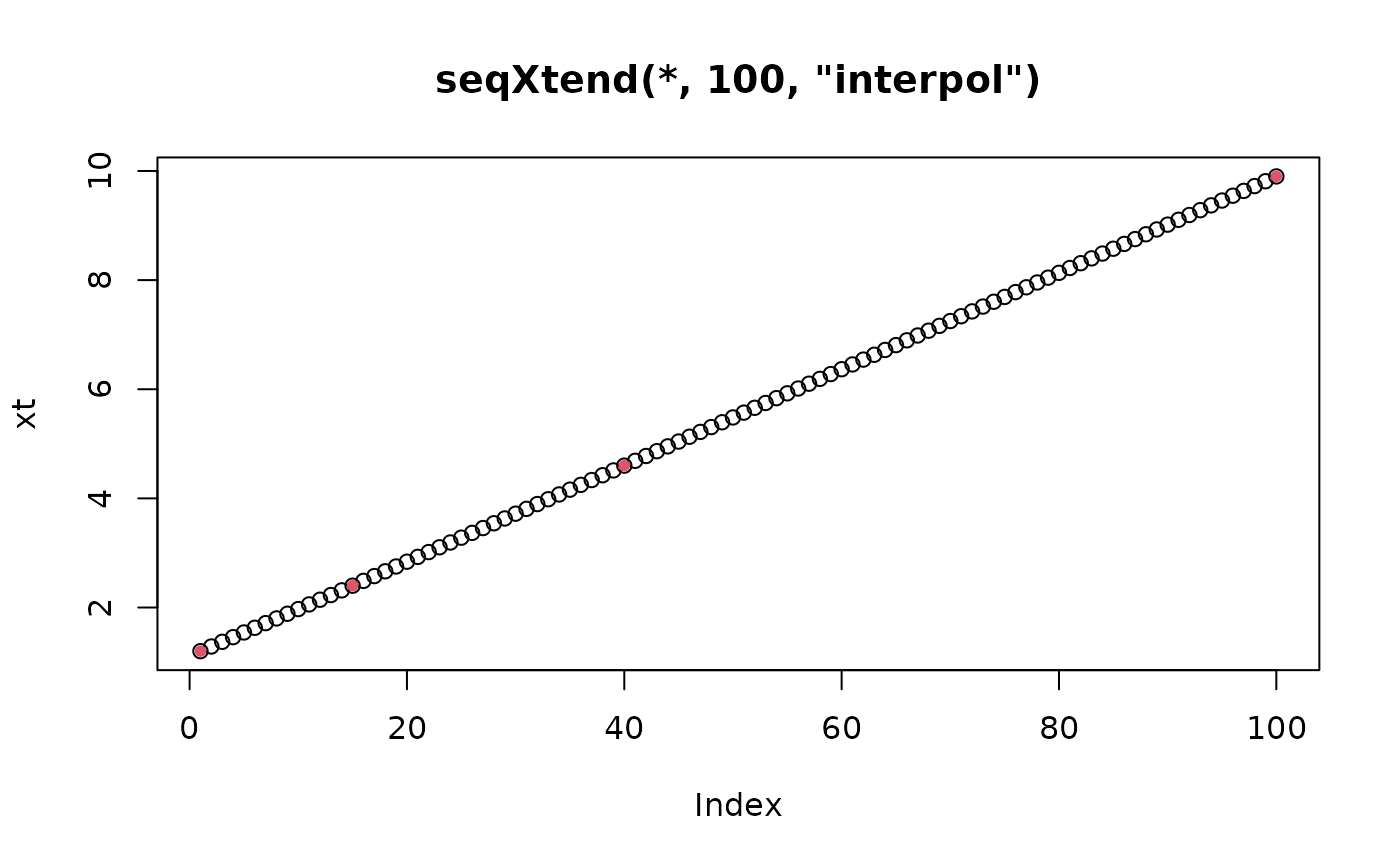 # .... you don't even see that it's not equidistant
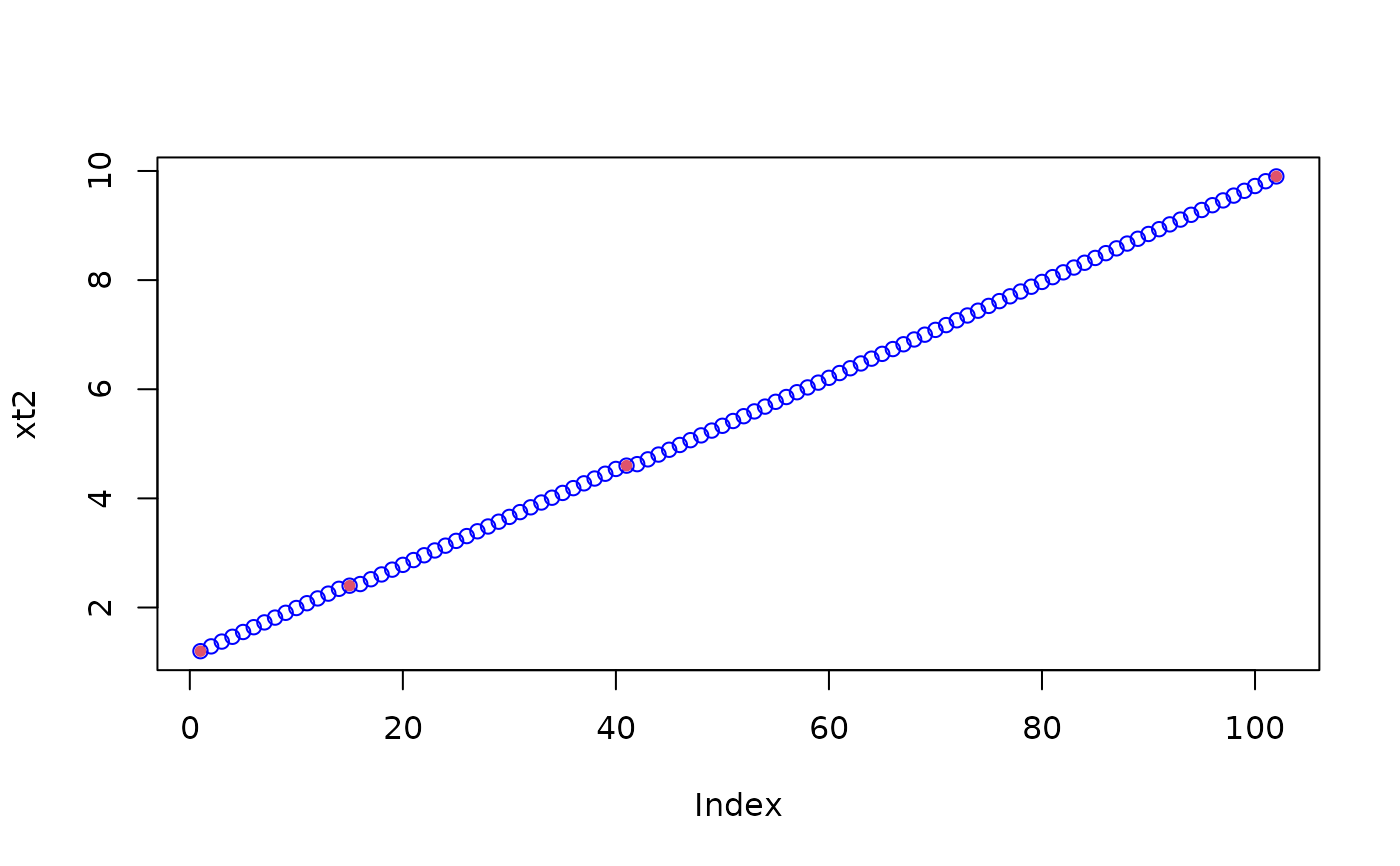
# whereas the cheap method shows ...
xt2 <- seqXtend(x, 100)
plot(xt2, col="blue")
points(match(x,xt2), x, col = 2, pch = 20)
# .... you don't even see that it's not equidistant
# whereas the cheap method shows ...
xt2 <- seqXtend(x, 100)
plot(xt2, col="blue")
points(match(x,xt2), x, col = 2, pch = 20)
 ## with "Date" objects
Drng <- as.Date(c("2007-11-10", "2012-07-12"))
(px <- pretty(Drng, n = 16)) # say, for the main labels
#> [1] "2007-10-01" "2008-01-01" "2008-04-01" "2008-07-01" "2008-10-01"
#> [6] "2009-01-01" "2009-04-01" "2009-07-01" "2009-10-01" "2010-01-01"
#> [11] "2010-04-01" "2010-07-01" "2010-10-01" "2011-01-01" "2011-04-01"
#> [16] "2011-07-01" "2011-10-01" "2012-01-01" "2012-04-01" "2012-07-01"
#> [21] "2012-10-01"
## say, a finer grid, for ticks -- should be almost equidistant
n3 <- 3*length(px)
summary(as.numeric(diff(seqXtend(px, n3)))) # wildly varying
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 0.5 16.0 29.5 22.6 29.5 29.5
summary(as.numeric(diff(seqXtend(px, n3, "aim")))) # (ditto)
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 1.7 17.1 33.8 29.5 42.5 42.5
summary(as.numeric(diff(seqXtend(px, n3, "int")))) # around 30
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 23.0 30.0 30.3 29.5 30.7 30.7
## with "Date" objects
Drng <- as.Date(c("2007-11-10", "2012-07-12"))
(px <- pretty(Drng, n = 16)) # say, for the main labels
#> [1] "2007-10-01" "2008-01-01" "2008-04-01" "2008-07-01" "2008-10-01"
#> [6] "2009-01-01" "2009-04-01" "2009-07-01" "2009-10-01" "2010-01-01"
#> [11] "2010-04-01" "2010-07-01" "2010-10-01" "2011-01-01" "2011-04-01"
#> [16] "2011-07-01" "2011-10-01" "2012-01-01" "2012-04-01" "2012-07-01"
#> [21] "2012-10-01"
## say, a finer grid, for ticks -- should be almost equidistant
n3 <- 3*length(px)
summary(as.numeric(diff(seqXtend(px, n3)))) # wildly varying
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 0.5 16.0 29.5 22.6 29.5 29.5
summary(as.numeric(diff(seqXtend(px, n3, "aim")))) # (ditto)
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 1.7 17.1 33.8 29.5 42.5 42.5
summary(as.numeric(diff(seqXtend(px, n3, "int")))) # around 30
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 23.0 30.0 30.3 29.5 30.7 30.7