Add Lines or Points to a Survival Plot
lines.survfit.RdOften used to add the expected survival curve(s) to a Kaplan-Meier plot
generated with plot.survfit.
# S3 method for class 'survfit'
lines(x, type="s", pch=3, col=1, lty=1,
lwd=1, cex=1, mark.time=FALSE, xmax,
fun, conf.int=FALSE,
conf.times, conf.cap=.005, conf.offset=.012,
conf.type = c("log", "log-log", "plain", "logit", "arcsin"),
mark, noplot="(s0)", cumhaz= FALSE, cumprob= FALSE, ...)
# S3 method for class 'survexp'
lines(x, type="l", ...)
# S3 method for class 'survfit'
points(x, fun, censor=FALSE, col=1, pch,
noplot="(s0)", cumhaz=FALSE, ...)Arguments
- x
a survival object, generated from the
survfitorsurvexpfunctions.- type
the line type, as described in
lines. The default is a step function forsurvfitobjects, and a connected line forsurvexpobjects. All other arguments forlines.survexpare identical to those forlines.survfit.- col, lty, lwd, cex
vectors giving the mark symbol, color, line type, line width and character size for the added curves. Of this set only color is applicable to
points.- pch
plotting characters for points, in the style of
matplot, i.e., either a single string of characters of which the first will be used for the first curve, etc; or a vector of characters or integers, one element per curve.- mark
a historical alias for
pch- censor
should censoring times be displayed for the
pointsfunction?- mark.time
controls the labeling of the curves. If
FALSE, no labeling is done. IfTRUE, then curves are marked at each censoring time. Ifmark.timeis a numeric vector, then curves are marked at the specified time points.- xmax
optional cutoff for the right hand of the curves.
- fun
an arbitrary function defining a transformation of the survival curve. For example
fun=logis an alternative way to draw a log-survival curve (but with the axis labeled with log(S) values). Four often used transformations can be specified with a character argument instead: "log" is the same as using thelog=Toption, "event" plots cumulative events (f(y) = 1-y), "cumhaz" plots the cumulative hazard function (f(y) = -log(y)) and "cloglog" creates a complimentary log-log survival plot (f(y) = log(-log(y))) along with log scale for the x-axis.- conf.int
if
TRUE, confidence bands for the curves are also plotted. If set to"only", then only the CI bands are plotted, and the curve itself is left off. This can be useful for fine control over the colors or line types of a plot.- conf.times
optional vector of times at which to place a confidence bar on the curve(s). If present, these will be used instead of confidence bands.
- conf.cap
width of the horizontal cap on top of the confidence bars; only used if conf.times is used. A value of 1 is the width of the plot region.
- conf.offset
the offset for confidence bars, when there are multiple curves on the plot. A value of 1 is the width of the plot region. If this is a single number then each curve's bars are offset by this amount from the prior curve's bars, if it is a vector the values are used directly.
- conf.type
One of
"plain","log"(the default),"log-log","logit", or"none". Only enough of the string to uniquely identify it is necessary. The first option causes confidence intervals not to be generated. The second causes the standard intervalscurve +- k *se(curve), where k is determined fromconf.int. The log option calculates intervals based on the cumulative hazard or log(survival). The log-log option bases the intervals on the log hazard or log(-log(survival)), and the logit option on log(survival/(1-survival)).- noplot
for multi-state models, curves with this label will not be plotted. The default corresponds to an unspecified state.
- cumhaz
plot the cumulative hazard, rather than the survival or probability in state.
- cumprob
for a multi-state curve, plot the probabilities in state 1, (state1 + state2), (state1 + state2 + state3), .... If
cumprobis an integer vector the totals will be in the order indicated.- ...
other graphical parameters
Value
a list with components x and y, containing the coordinates of the
last point on each of the curves (but not of the confidence limits).
This may be useful for labeling.
If cumprob=TRUE then y will be a matrix with one row per
curve and x will be all the time points. This may be useful for
adding shading.
Side Effects
one or more curves are added to the current plot.
See also
lines, par, plot.survfit, survfit, survexp.
Details
When the survfit function creates a multi-state survival curve
the resulting object has class `survfitms'. The only difference in
the plots is that that it defaults to a curve that goes from lower
left to upper right (starting at 0), where survival curves default
to starting at 1 and going down. All other options are identical.
If the user set an explicit range in an earlier plot.survfit
call, e.g. via xlim or xmax, subsequent calls to
this function remember the right hand cutoff. This memory can be
erased by options(plot.survfit) <- NULL.
Examples
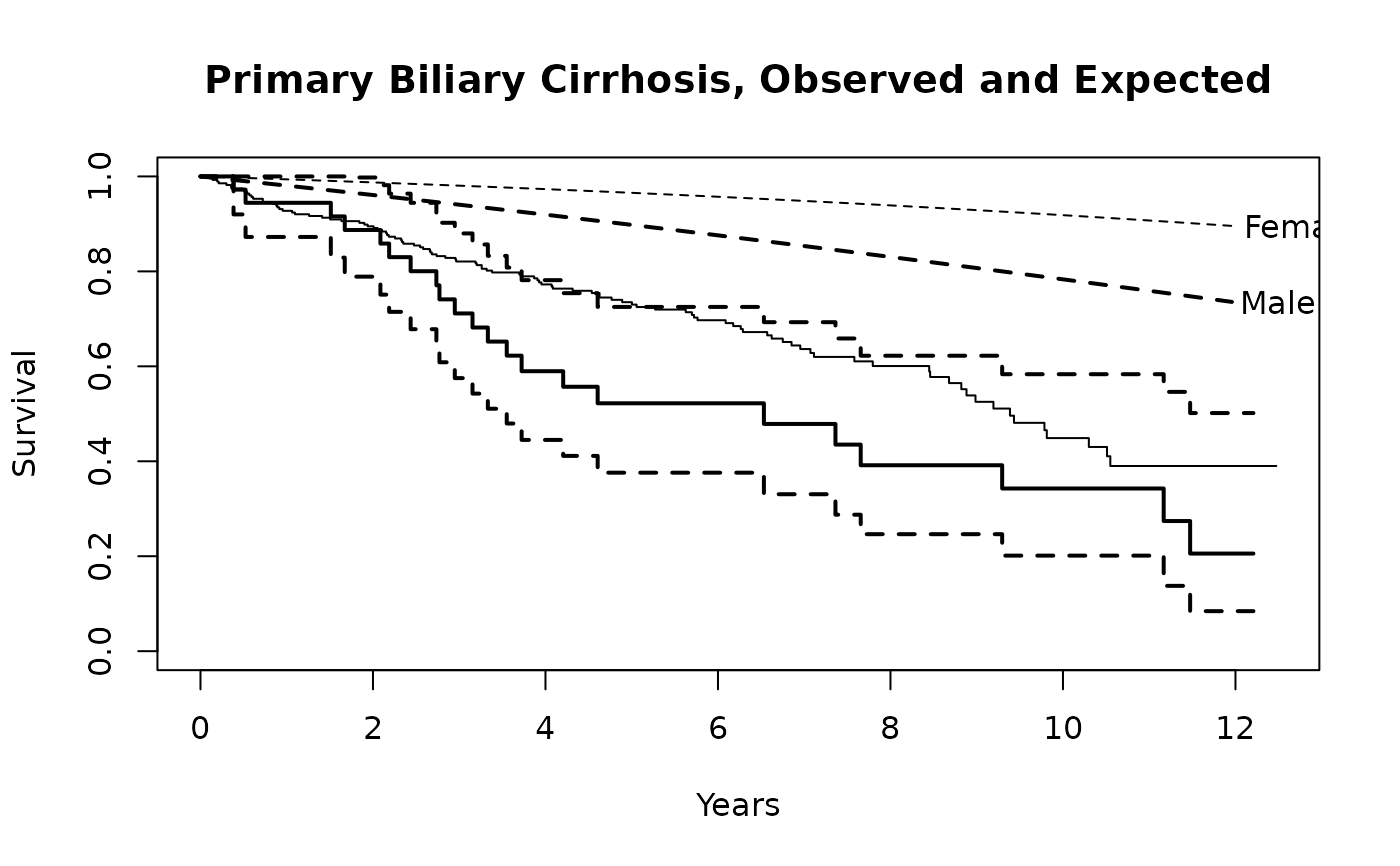
fit <- survfit(Surv(time, status==2) ~ sex, pbc,subset=1:312)
plot(fit, mark.time=FALSE, xscale=365.25,
xlab='Years', ylab='Survival')
lines(fit[1], lwd=2) #darken the first curve and add marks
# Add expected survival curves for the two groups,
# based on the US census data
# The data set does not have entry date, use the midpoint of the study
efit <- survexp(~sex, data=pbc, times= (0:24)*182, ratetable=survexp.us,
rmap=list(sex=sex, age=age*365.35, year=as.Date('1979/01/01')))
temp <- lines(efit, lty=2, lwd=2:1)
text(temp, c("Male", "Female"), adj= -.1) #labels just past the ends
title(main="Primary Biliary Cirrhosis, Observed and Expected")