Body Temperature Series of Beaver 2
beav2.RdReynolds (1994) describes a small part of a study of the long-term temperature dynamics of beaver Castor canadensis in north-central Wisconsin. Body temperature was measured by telemetry every 10 minutes for four females, but data from a one period of less than a day for each of two animals is used there.
beav2Format
The beav2 data frame has 100 rows and 4 columns.
This data frame contains the following columns:
dayDay of observation (in days since the beginning of 1990), November 3–4.
timeTime of observation, in the form
0330for 3.30am.tempMeasured body temperature in degrees Celsius.
activIndicator of activity outside the retreat.
Source
P. S. Reynolds (1994) Time-series analyses of beaver body temperatures. Chapter 11 of Lange, N., Ryan, L., Billard, L., Brillinger, D., Conquest, L. and Greenhouse, J. eds (1994) Case Studies in Biometry. New York: John Wiley and Sons.
References
Venables, W. N. and Ripley, B. D. (2002) Modern Applied Statistics with S. Fourth edition. Springer.
See also
Examples
attach(beav2)
beav2$hours <- 24*(day-307) + trunc(time/100) + (time%%100)/60
plot(beav2$hours, beav2$temp, type = "l", xlab = "time",
ylab = "temperature", main = "Beaver 2")
usr <- par("usr"); usr[3:4] <- c(-0.2, 8); par(usr = usr)
lines(beav2$hours, beav2$activ, type = "s", lty = 2)
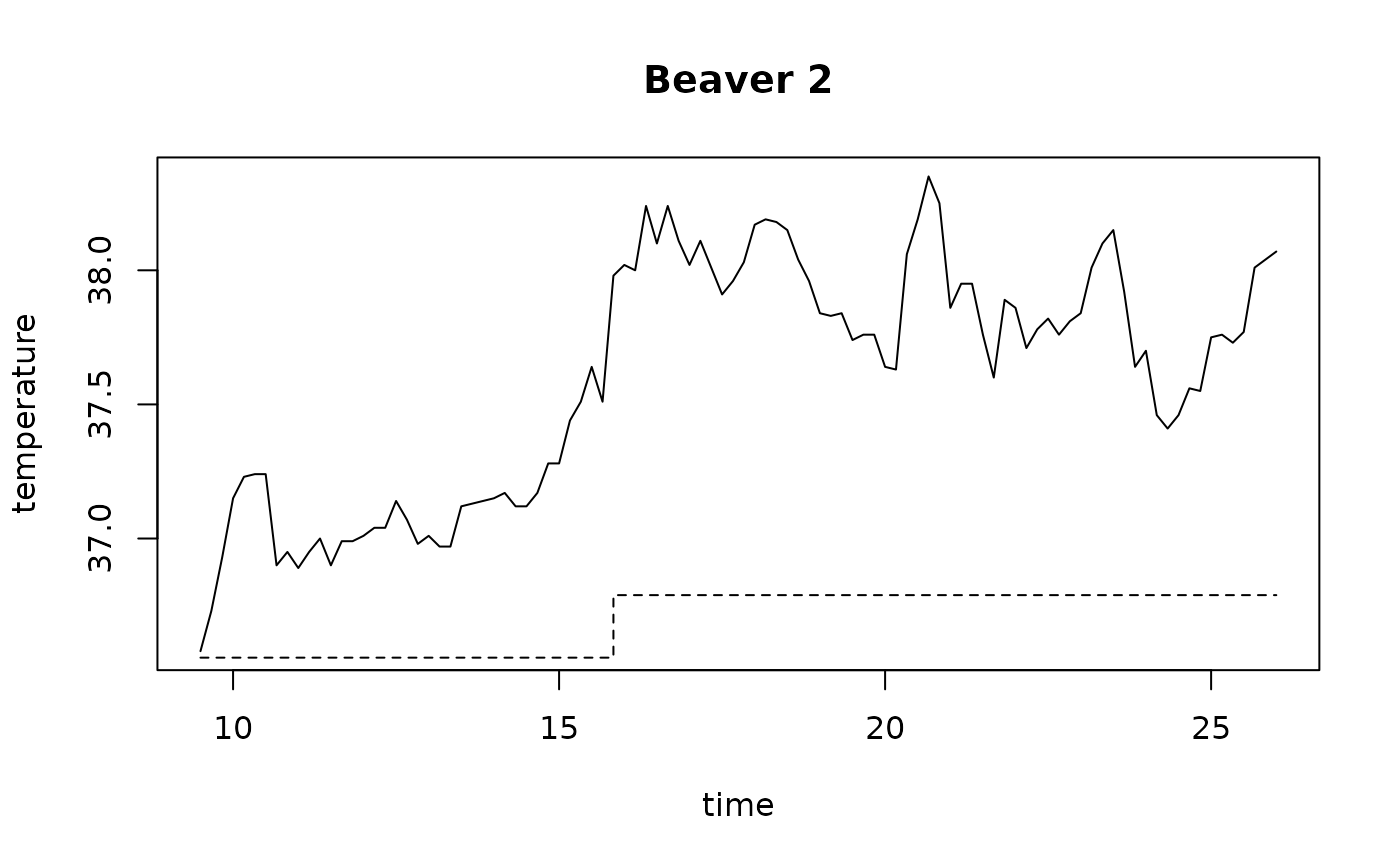 temp <- ts(temp, start = 8+2/3, frequency = 6)
activ <- ts(activ, start = 8+2/3, frequency = 6)
acf(temp[activ == 0]); acf(temp[activ == 1]) # also look at PACFs
temp <- ts(temp, start = 8+2/3, frequency = 6)
activ <- ts(activ, start = 8+2/3, frequency = 6)
acf(temp[activ == 0]); acf(temp[activ == 1]) # also look at PACFs
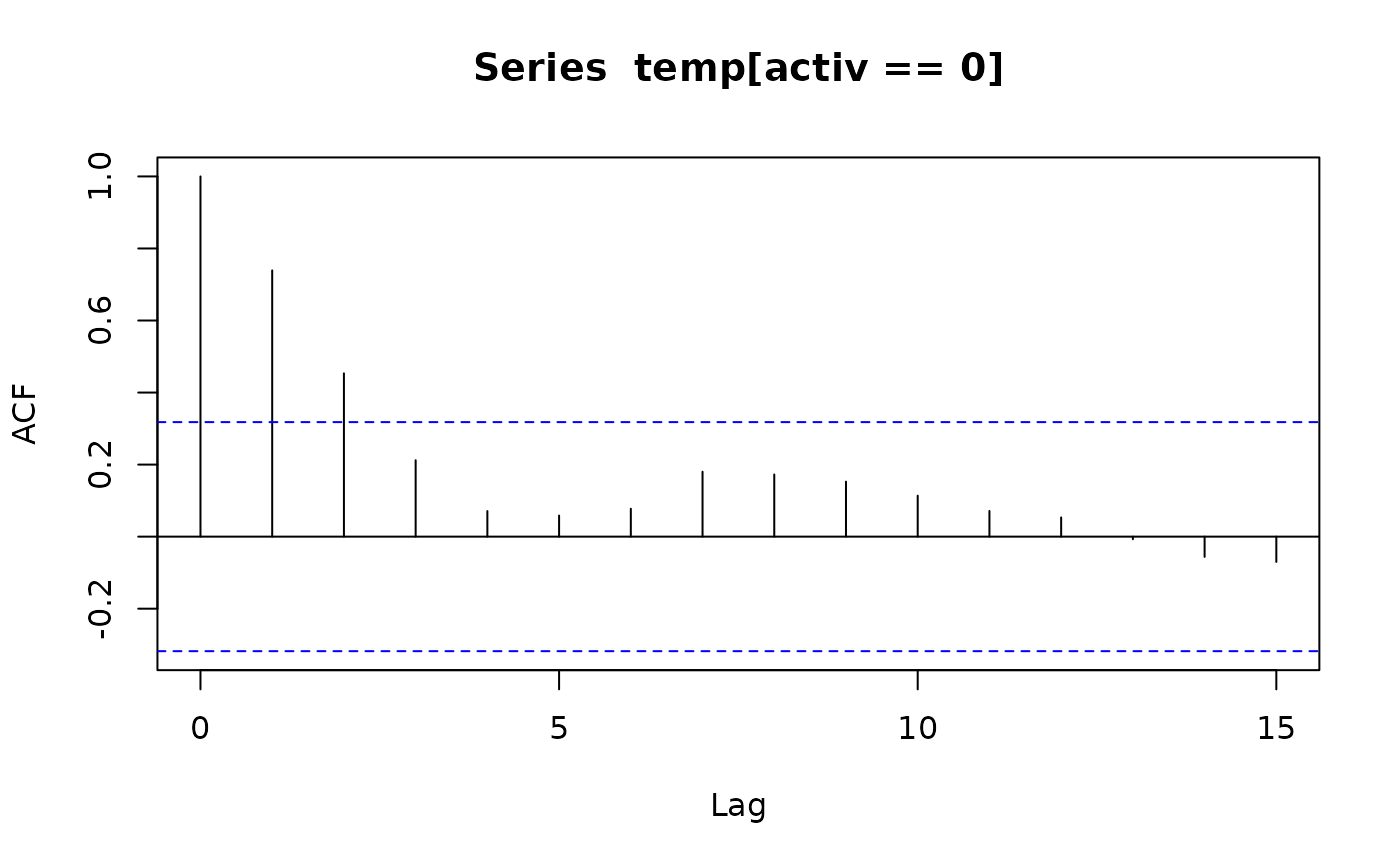
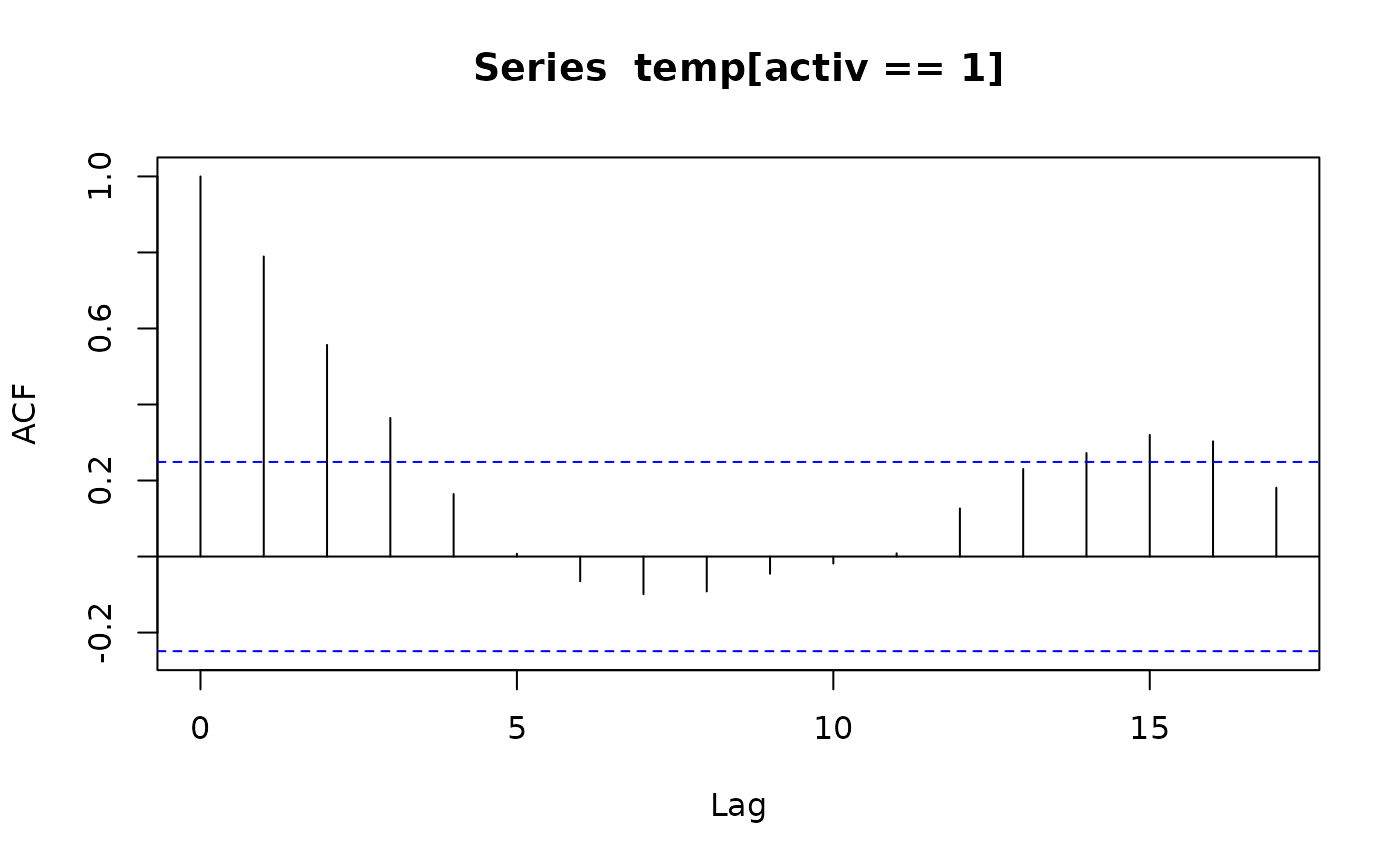 ar(temp[activ == 0]); ar(temp[activ == 1])
#>
#> Call:
#> ar(x = temp[activ == 0])
#>
#> Coefficients:
#> 1
#> 0.7392
#>
#> Order selected 1 sigma^2 estimated as 0.02011
#>
#> Call:
#> ar(x = temp[activ == 1])
#>
#> Coefficients:
#> 1
#> 0.7894
#>
#> Order selected 1 sigma^2 estimated as 0.01792
arima(temp, order = c(1,0,0), xreg = activ)
#>
#> Call:
#> arima(x = temp, order = c(1, 0, 0), xreg = activ)
#>
#> Coefficients:
#> ar1 intercept activ
#> 0.8733 37.1920 0.6139
#> s.e. 0.0684 0.1187 0.1381
#>
#> sigma^2 estimated as 0.01518: log likelihood = 66.78, aic = -125.55
dreg <- cbind(sin = sin(2*pi*beav2$hours/24), cos = cos(2*pi*beav2$hours/24))
arima(temp, order = c(1,0,0), xreg = cbind(active=activ, dreg))
#>
#> Call:
#> arima(x = temp, order = c(1, 0, 0), xreg = cbind(active = activ, dreg))
#>
#> Coefficients:
#> ar1 intercept active dreg.sin dreg.cos
#> 0.7905 37.1674 0.5322 -0.282 0.1201
#> s.e. 0.0681 0.0939 0.1282 0.105 0.0997
#>
#> sigma^2 estimated as 0.01434: log likelihood = 69.83, aic = -127.67
## IGNORE_RDIFF_BEGIN
library(nlme) # for gls and corAR1
beav2.gls <- gls(temp ~ activ, data = beav2, correlation = corAR1(0.8),
method = "ML")
summary(beav2.gls)
#> Generalized least squares fit by maximum likelihood
#> Model: temp ~ activ
#> Data: beav2
#> AIC BIC logLik
#> -125.5505 -115.1298 66.77523
#>
#> Correlation Structure: AR(1)
#> Formula: ~1
#> Parameter estimate(s):
#> Phi
#> 0.8731771
#>
#> Coefficients:
#> Value Std.Error t-value p-value
#> (Intercept) 37.19195 0.1131328 328.7460 0
#> activ 0.61418 0.1087286 5.6487 0
#>
#> Correlation:
#> (Intr)
#> activ -0.582
#>
#> Standardized residuals:
#> Min Q1 Med Q3 Max
#> -2.42080776 -0.61510520 -0.03573836 0.81641138 2.15153496
#>
#> Residual standard error: 0.2527856
#> Degrees of freedom: 100 total; 98 residual
summary(update(beav2.gls, subset = 6:100))
#> Generalized least squares fit by maximum likelihood
#> Model: temp ~ activ
#> Data: beav2
#> Subset: 6:100
#> AIC BIC logLik
#> -124.981 -114.7654 66.49048
#>
#> Correlation Structure: AR(1)
#> Formula: ~1
#> Parameter estimate(s):
#> Phi
#> 0.8380448
#>
#> Coefficients:
#> Value Std.Error t-value p-value
#> (Intercept) 37.25001 0.09634047 386.6496 0
#> activ 0.60277 0.09931904 6.0690 0
#>
#> Correlation:
#> (Intr)
#> activ -0.657
#>
#> Standardized residuals:
#> Min Q1 Med Q3 Max
#> -2.0231494 -0.8910348 -0.1497564 0.7640939 2.2719468
#>
#> Residual standard error: 0.2188542
#> Degrees of freedom: 95 total; 93 residual
detach("beav2"); rm(temp, activ)
## IGNORE_RDIFF_END
ar(temp[activ == 0]); ar(temp[activ == 1])
#>
#> Call:
#> ar(x = temp[activ == 0])
#>
#> Coefficients:
#> 1
#> 0.7392
#>
#> Order selected 1 sigma^2 estimated as 0.02011
#>
#> Call:
#> ar(x = temp[activ == 1])
#>
#> Coefficients:
#> 1
#> 0.7894
#>
#> Order selected 1 sigma^2 estimated as 0.01792
arima(temp, order = c(1,0,0), xreg = activ)
#>
#> Call:
#> arima(x = temp, order = c(1, 0, 0), xreg = activ)
#>
#> Coefficients:
#> ar1 intercept activ
#> 0.8733 37.1920 0.6139
#> s.e. 0.0684 0.1187 0.1381
#>
#> sigma^2 estimated as 0.01518: log likelihood = 66.78, aic = -125.55
dreg <- cbind(sin = sin(2*pi*beav2$hours/24), cos = cos(2*pi*beav2$hours/24))
arima(temp, order = c(1,0,0), xreg = cbind(active=activ, dreg))
#>
#> Call:
#> arima(x = temp, order = c(1, 0, 0), xreg = cbind(active = activ, dreg))
#>
#> Coefficients:
#> ar1 intercept active dreg.sin dreg.cos
#> 0.7905 37.1674 0.5322 -0.282 0.1201
#> s.e. 0.0681 0.0939 0.1282 0.105 0.0997
#>
#> sigma^2 estimated as 0.01434: log likelihood = 69.83, aic = -127.67
## IGNORE_RDIFF_BEGIN
library(nlme) # for gls and corAR1
beav2.gls <- gls(temp ~ activ, data = beav2, correlation = corAR1(0.8),
method = "ML")
summary(beav2.gls)
#> Generalized least squares fit by maximum likelihood
#> Model: temp ~ activ
#> Data: beav2
#> AIC BIC logLik
#> -125.5505 -115.1298 66.77523
#>
#> Correlation Structure: AR(1)
#> Formula: ~1
#> Parameter estimate(s):
#> Phi
#> 0.8731771
#>
#> Coefficients:
#> Value Std.Error t-value p-value
#> (Intercept) 37.19195 0.1131328 328.7460 0
#> activ 0.61418 0.1087286 5.6487 0
#>
#> Correlation:
#> (Intr)
#> activ -0.582
#>
#> Standardized residuals:
#> Min Q1 Med Q3 Max
#> -2.42080776 -0.61510520 -0.03573836 0.81641138 2.15153496
#>
#> Residual standard error: 0.2527856
#> Degrees of freedom: 100 total; 98 residual
summary(update(beav2.gls, subset = 6:100))
#> Generalized least squares fit by maximum likelihood
#> Model: temp ~ activ
#> Data: beav2
#> Subset: 6:100
#> AIC BIC logLik
#> -124.981 -114.7654 66.49048
#>
#> Correlation Structure: AR(1)
#> Formula: ~1
#> Parameter estimate(s):
#> Phi
#> 0.8380448
#>
#> Coefficients:
#> Value Std.Error t-value p-value
#> (Intercept) 37.25001 0.09634047 386.6496 0
#> activ 0.60277 0.09931904 6.0690 0
#>
#> Correlation:
#> (Intr)
#> activ -0.657
#>
#> Standardized residuals:
#> Min Q1 Med Q3 Max
#> -2.0231494 -0.8910348 -0.1497564 0.7640939 2.2719468
#>
#> Residual standard error: 0.2188542
#> Degrees of freedom: 95 total; 93 residual
detach("beav2"); rm(temp, activ)
## IGNORE_RDIFF_END