Fixing a Quantile Regression having Crossing
fix.crossing.RdReturns a similar object fitted with columns of the constraint matrices amalgamated so it is a partially parallel VGLM object. The columns combined correspond to certain crossing quantiles. This applies especially to an extlogF1() VGLM object.
fix.crossing.vglm(object, maxit = 100, trace = FALSE, ...)Arguments
- object
an object such as a
vglmobject with family functionextlogF1.- maxit, trace
values for overwriting components in
vglm.control. Setting these toNULLwill mean the values invglm.controlonobjectwill be retained.- ...
additional optional arguments. Currently unused.
Details
The quantile crossing problem has been described as
disturbing and embarrassing.
This function was specifically written for
a vglm with family function extlogF1.
It examines the fitted quantiles of object to see if any cross.
If so, then a pair of columns is combined to make those
two quantiles parallel.
After fitting the submodel it then repeats testing for
crossing quantiles and repairing them, until there is
no more quantile crossing detected.
Note that it is possible that the quantiles cross in
some subset of the covariate space not covered by the
data—see is.crossing.
This function is fragile and likely to change in the future.
For extlogF1 models, it is assumed
that argument data has been assigned a data frame,
and
that the default values of the argument parallel
has been used; this means that the second constraint
matrix is diag(M).
The constraint matrix of the intercept term remains unchanged
as diag(M).
Value
An object very similar to the original object, but with possibly different constraint matrices (partially parallel) so as to remove any quantile crossing.
See also
Examples
if (FALSE) ooo <- with(bmi.nz, order(age))
bmi.nz <- bmi.nz[ooo, ] # Sort by age
#> Error: object 'ooo' not found
with(bmi.nz, plot(age, BMI, col = "blue"))
 mytau <- c(50, 93, 95, 97) / 100 # Some quantiles are quite close
fit1 <- vglm(BMI ~ ns(age, 7), extlogF1(mytau), bmi.nz, trace = TRUE)
#> Taking a modified step.
#> Iteration 2 : loglikelihood = -9827.2672
#> Taking a modified step.
#> Iteration 3 : loglikelihood = -9821.8497
#> Taking a modified step.
#> Iteration 4 : loglikelihood = -9817.6926
#> Taking a modified step.
#> Iteration 5 : loglikelihood = -9814.6062
#> Taking a modified step.
#> Iteration 6 : loglikelihood = -9812.2572
#> Taking a modified step.
#> Iteration 7 : loglikelihood = -9810.5167
#> Taking a modified step.
#> Iteration 8 : loglikelihood = -9809.2165
#> Taking a modified step.
#> Iteration 9 : loglikelihood = -9808.2009
#> Taking a modified step.
#> Iteration 10 : loglikelihood = -9807.4706
#> Taking a modified step.
#> Iteration 11 : loglikelihood = -9807.0344
#> Taking a modified step.
#> Iteration 12 : loglikelihood = -9806.7858
#> Taking a modified step.
#> Iteration 13 : loglikelihood = -9806.6309
#> Taking a modified step.
#> Iteration 14 : loglikelihood = -9806.5045
#> Taking a modified step.
#> Iteration 15 : loglikelihood = -9806.3928
#> Taking a modified step.
#> Iteration 16 : loglikelihood = -9806.2922
#> Taking a modified step.
#> Iteration 17 : loglikelihood = -9806.2011
#> Taking a modified step.
#> Iteration 18 : loglikelihood = -9806.1193
#> Taking a modified step.
#> Iteration 19 : loglikelihood = -9806.047
#> Taking a modified step.
#> Iteration 20 : loglikelihood = -9805.9853
#> Taking a modified step.
#> Iteration 21 : loglikelihood = -9805.935
#> Taking a modified step.
#> Iteration 22 : loglikelihood = -9805.8964
#> Taking a modified step.
#> Iteration 23 : loglikelihood = -9805.8694
#> Taking a modified step.
#> Iteration 24 : loglikelihood = -9805.8525
#> Taking a modified step.
#> Iteration 25 : loglikelihood = -9805.8431
#> Taking a modified step.
#> Iteration 26 : loglikelihood = -9805.8382
#> Taking a modified step.
#> Iteration 27 : loglikelihood = -9805.8355
#> Taking a modified step.
#> Iteration 28 : loglikelihood = -9805.8338
#> Taking a modified step.
#> Iteration 29 : loglikelihood = -9805.8326
#> Taking a modified step.
#> Iteration 30 : loglikelihood = -9805.8316
#> Taking a modified step.
#> Iteration 31 : loglikelihood = -9805.8309
#> Taking a modified step.
#> Iteration 32 : loglikelihood = -9805.8304
#> Taking a modified step.
#> Iteration 33 : loglikelihood = -9805.83
#> Taking a modified step.
#> Iteration 34 : loglikelihood = -9805.8297
#> Taking a modified step.
#> Iteration 35 : loglikelihood = -9805.8295
#> Taking a modified step.
#> Iteration 36 : loglikelihood = -9805.8293
#> Taking a modified step.
#> Iteration 37 : loglikelihood = -9805.8291
#> Taking a modified step.
#> Iteration 38 : loglikelihood = -9805.829
#> Taking a modified step.
#> Iteration 39 : loglikelihood = -9805.8289
#> Taking a modified step.
#> Iteration 40 : loglikelihood = -9805.8289
#> Taking a modified step.
#> Iteration 41 : loglikelihood = -9805.8288
#> Taking a modified step.
#> Iteration 42 : loglikelihood = -9805.8288
#> Taking a modified step.
#> Iteration 43 : loglikelihood = -9805.8288
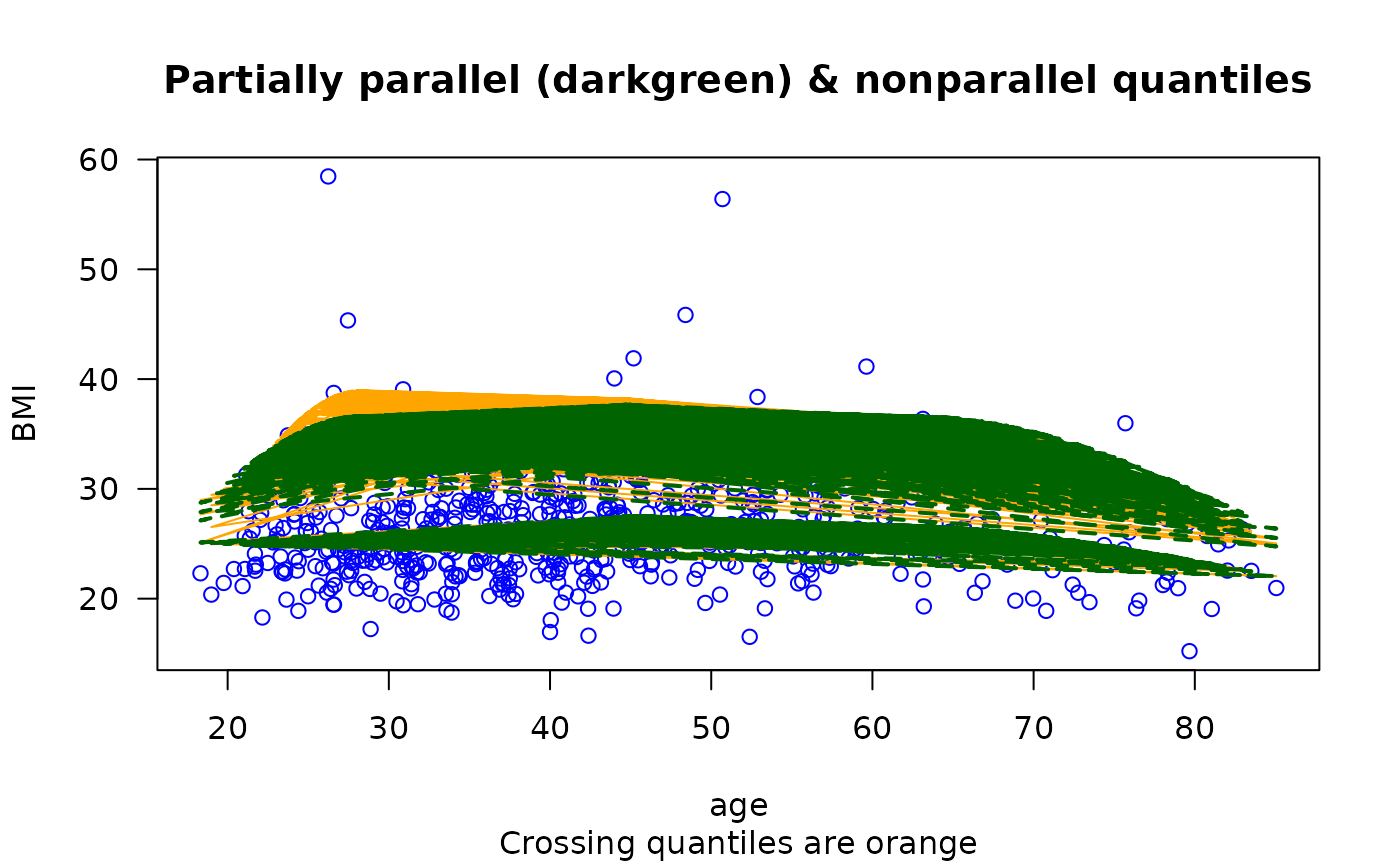
plot(BMI ~ age, bmi.nz, col = "blue", las = 1,
main = "Partially parallel (darkgreen) & nonparallel quantiles",
sub = "Crossing quantiles are orange")
fix.crossing(fit1)
#>
#> Call:
#> vglm(formula = as.formula(formula(object)), family = extlogF1(tau = object.save@extra$tau,
#> lambda.arg = object.save@extra$lambda.arg, scale.arg = object.save@extra$scale.arg,
#> llocation = linkfun(object.save)[1], parallel = FALSE), data = get(object.save@misc$dataname),
#> control = object@control, constraints = Hlist)
#>
#>
#> Coefficients:
#> (Intercept):1 (Intercept):2 (Intercept):3 (Intercept):4 ns(age, 7)1:1
#> 25.1146001 27.1231849 27.8923265 28.7466503 1.0290005
#> ns(age, 7)1:2 ns(age, 7)2:1 ns(age, 7)2:2 ns(age, 7)3:1 ns(age, 7)3:2
#> 4.8016099 0.1720977 5.0590385 3.3312391 11.0238041
#> ns(age, 7)4:1 ns(age, 7)4:2 ns(age, 7)5:1 ns(age, 7)5:2 ns(age, 7)6:1
#> 0.7732957 3.9197381 2.8414896 7.4504449 -1.2498084
#> ns(age, 7)6:2 ns(age, 7)7:1 ns(age, 7)7:2
#> 12.6545997 -2.7057898 -7.5027877
#>
#> Degrees of Freedom: 2800 Total; 2782 Residual
#> Log-likelihood: -9808.988
#>
#> Quantiles:
#> (tau = 0.5) (tau = 0.93) (tau = 0.95) (tau = 0.97)
#> 0.5028571 0.9257143 0.9485714 0.9742857
matlines(with(bmi.nz, age), fitted(fit1), lty = 1, col = "orange")
fit2 <- fix.crossing(fit1) # Some quantiles have been fixed
constraints(fit2)
#> $`(Intercept)`
#> [,1] [,2] [,3] [,4]
#> [1,] 1 0 0 0
#> [2,] 0 1 0 0
#> [3,] 0 0 1 0
#> [4,] 0 0 0 1
#>
#> $`ns(age, 7)1`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
#> $`ns(age, 7)2`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
#> $`ns(age, 7)3`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
#> $`ns(age, 7)4`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
#> $`ns(age, 7)5`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
#> $`ns(age, 7)6`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
#> $`ns(age, 7)7`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
matlines(with(bmi.nz, age), fitted(fit2), lty = "dashed",
col = "darkgreen", lwd = 2) # \dontrun{}
mytau <- c(50, 93, 95, 97) / 100 # Some quantiles are quite close
fit1 <- vglm(BMI ~ ns(age, 7), extlogF1(mytau), bmi.nz, trace = TRUE)
#> Taking a modified step.
#> Iteration 2 : loglikelihood = -9827.2672
#> Taking a modified step.
#> Iteration 3 : loglikelihood = -9821.8497
#> Taking a modified step.
#> Iteration 4 : loglikelihood = -9817.6926
#> Taking a modified step.
#> Iteration 5 : loglikelihood = -9814.6062
#> Taking a modified step.
#> Iteration 6 : loglikelihood = -9812.2572
#> Taking a modified step.
#> Iteration 7 : loglikelihood = -9810.5167
#> Taking a modified step.
#> Iteration 8 : loglikelihood = -9809.2165
#> Taking a modified step.
#> Iteration 9 : loglikelihood = -9808.2009
#> Taking a modified step.
#> Iteration 10 : loglikelihood = -9807.4706
#> Taking a modified step.
#> Iteration 11 : loglikelihood = -9807.0344
#> Taking a modified step.
#> Iteration 12 : loglikelihood = -9806.7858
#> Taking a modified step.
#> Iteration 13 : loglikelihood = -9806.6309
#> Taking a modified step.
#> Iteration 14 : loglikelihood = -9806.5045
#> Taking a modified step.
#> Iteration 15 : loglikelihood = -9806.3928
#> Taking a modified step.
#> Iteration 16 : loglikelihood = -9806.2922
#> Taking a modified step.
#> Iteration 17 : loglikelihood = -9806.2011
#> Taking a modified step.
#> Iteration 18 : loglikelihood = -9806.1193
#> Taking a modified step.
#> Iteration 19 : loglikelihood = -9806.047
#> Taking a modified step.
#> Iteration 20 : loglikelihood = -9805.9853
#> Taking a modified step.
#> Iteration 21 : loglikelihood = -9805.935
#> Taking a modified step.
#> Iteration 22 : loglikelihood = -9805.8964
#> Taking a modified step.
#> Iteration 23 : loglikelihood = -9805.8694
#> Taking a modified step.
#> Iteration 24 : loglikelihood = -9805.8525
#> Taking a modified step.
#> Iteration 25 : loglikelihood = -9805.8431
#> Taking a modified step.
#> Iteration 26 : loglikelihood = -9805.8382
#> Taking a modified step.
#> Iteration 27 : loglikelihood = -9805.8355
#> Taking a modified step.
#> Iteration 28 : loglikelihood = -9805.8338
#> Taking a modified step.
#> Iteration 29 : loglikelihood = -9805.8326
#> Taking a modified step.
#> Iteration 30 : loglikelihood = -9805.8316
#> Taking a modified step.
#> Iteration 31 : loglikelihood = -9805.8309
#> Taking a modified step.
#> Iteration 32 : loglikelihood = -9805.8304
#> Taking a modified step.
#> Iteration 33 : loglikelihood = -9805.83
#> Taking a modified step.
#> Iteration 34 : loglikelihood = -9805.8297
#> Taking a modified step.
#> Iteration 35 : loglikelihood = -9805.8295
#> Taking a modified step.
#> Iteration 36 : loglikelihood = -9805.8293
#> Taking a modified step.
#> Iteration 37 : loglikelihood = -9805.8291
#> Taking a modified step.
#> Iteration 38 : loglikelihood = -9805.829
#> Taking a modified step.
#> Iteration 39 : loglikelihood = -9805.8289
#> Taking a modified step.
#> Iteration 40 : loglikelihood = -9805.8289
#> Taking a modified step.
#> Iteration 41 : loglikelihood = -9805.8288
#> Taking a modified step.
#> Iteration 42 : loglikelihood = -9805.8288
#> Taking a modified step.
#> Iteration 43 : loglikelihood = -9805.8288
plot(BMI ~ age, bmi.nz, col = "blue", las = 1,
main = "Partially parallel (darkgreen) & nonparallel quantiles",
sub = "Crossing quantiles are orange")
fix.crossing(fit1)
#>
#> Call:
#> vglm(formula = as.formula(formula(object)), family = extlogF1(tau = object.save@extra$tau,
#> lambda.arg = object.save@extra$lambda.arg, scale.arg = object.save@extra$scale.arg,
#> llocation = linkfun(object.save)[1], parallel = FALSE), data = get(object.save@misc$dataname),
#> control = object@control, constraints = Hlist)
#>
#>
#> Coefficients:
#> (Intercept):1 (Intercept):2 (Intercept):3 (Intercept):4 ns(age, 7)1:1
#> 25.1146001 27.1231849 27.8923265 28.7466503 1.0290005
#> ns(age, 7)1:2 ns(age, 7)2:1 ns(age, 7)2:2 ns(age, 7)3:1 ns(age, 7)3:2
#> 4.8016099 0.1720977 5.0590385 3.3312391 11.0238041
#> ns(age, 7)4:1 ns(age, 7)4:2 ns(age, 7)5:1 ns(age, 7)5:2 ns(age, 7)6:1
#> 0.7732957 3.9197381 2.8414896 7.4504449 -1.2498084
#> ns(age, 7)6:2 ns(age, 7)7:1 ns(age, 7)7:2
#> 12.6545997 -2.7057898 -7.5027877
#>
#> Degrees of Freedom: 2800 Total; 2782 Residual
#> Log-likelihood: -9808.988
#>
#> Quantiles:
#> (tau = 0.5) (tau = 0.93) (tau = 0.95) (tau = 0.97)
#> 0.5028571 0.9257143 0.9485714 0.9742857
matlines(with(bmi.nz, age), fitted(fit1), lty = 1, col = "orange")
fit2 <- fix.crossing(fit1) # Some quantiles have been fixed
constraints(fit2)
#> $`(Intercept)`
#> [,1] [,2] [,3] [,4]
#> [1,] 1 0 0 0
#> [2,] 0 1 0 0
#> [3,] 0 0 1 0
#> [4,] 0 0 0 1
#>
#> $`ns(age, 7)1`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
#> $`ns(age, 7)2`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
#> $`ns(age, 7)3`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
#> $`ns(age, 7)4`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
#> $`ns(age, 7)5`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
#> $`ns(age, 7)6`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
#> $`ns(age, 7)7`
#> [,1] [,2]
#> [1,] 1 0
#> [2,] 0 1
#> [3,] 0 1
#> [4,] 0 1
#>
matlines(with(bmi.nz, age), fitted(fit2), lty = "dashed",
col = "darkgreen", lwd = 2) # \dontrun{}