Quantile Crossing Detection
is.crossing.RdReturns a logical from testing whether an object such as an extlogF1() VGLM object has crossing quantiles.
is.crossing.vglm(object, ...)Arguments
Details
This function was specifically written for
a vglm with family function extlogF1.
It examines the fitted quantiles to see if any cross.
Note that if one uses regression splines such as
bs and
ns
then it is possible that they cross at values of the
covariate space that are not represented by actual data.
One could use linear interpolation between fitted values
to get around this problem.
Value
A logical.
If TRUE then one can try fit a similar model by
combining columns of the constraint matrices so that
crossing no longer holds; see fix.crossing.
For LMS-Box-Cox type quantile regression models
it is impossible for the quantiles to cross, by definition,
hence FALSE is returned;
see lms.bcn.
See also
Examples
if (FALSE) ooo <- with(bmi.nz, order(age))
bmi.nz <- bmi.nz[ooo, ] # Sort by age
#> Error: object 'ooo' not found
with(bmi.nz, plot(age, BMI, col = "blue"))
 mytau <- c(50, 93, 95, 97) / 100 # Some quantiles are quite close
fit1 <- vglm(BMI ~ ns(age, 7), extlogF1(mytau), bmi.nz, trace = TRUE)
#> Taking a modified step.
#> Iteration 2 : loglikelihood = -9827.2672
#> Taking a modified step.
#> Iteration 3 : loglikelihood = -9821.8497
#> Taking a modified step.
#> Iteration 4 : loglikelihood = -9817.6926
#> Taking a modified step.
#> Iteration 5 : loglikelihood = -9814.6062
#> Taking a modified step.
#> Iteration 6 : loglikelihood = -9812.2572
#> Taking a modified step.
#> Iteration 7 : loglikelihood = -9810.5167
#> Taking a modified step.
#> Iteration 8 : loglikelihood = -9809.2165
#> Taking a modified step.
#> Iteration 9 : loglikelihood = -9808.2009
#> Taking a modified step.
#> Iteration 10 : loglikelihood = -9807.4706
#> Taking a modified step.
#> Iteration 11 : loglikelihood = -9807.0344
#> Taking a modified step.
#> Iteration 12 : loglikelihood = -9806.7858
#> Taking a modified step.
#> Iteration 13 : loglikelihood = -9806.6309
#> Taking a modified step.
#> Iteration 14 : loglikelihood = -9806.5045
#> Taking a modified step.
#> Iteration 15 : loglikelihood = -9806.3928
#> Taking a modified step.
#> Iteration 16 : loglikelihood = -9806.2922
#> Taking a modified step.
#> Iteration 17 : loglikelihood = -9806.2011
#> Taking a modified step.
#> Iteration 18 : loglikelihood = -9806.1193
#> Taking a modified step.
#> Iteration 19 : loglikelihood = -9806.047
#> Taking a modified step.
#> Iteration 20 : loglikelihood = -9805.9853
#> Taking a modified step.
#> Iteration 21 : loglikelihood = -9805.935
#> Taking a modified step.
#> Iteration 22 : loglikelihood = -9805.8964
#> Taking a modified step.
#> Iteration 23 : loglikelihood = -9805.8694
#> Taking a modified step.
#> Iteration 24 : loglikelihood = -9805.8525
#> Taking a modified step.
#> Iteration 25 : loglikelihood = -9805.8431
#> Taking a modified step.
#> Iteration 26 : loglikelihood = -9805.8382
#> Taking a modified step.
#> Iteration 27 : loglikelihood = -9805.8355
#> Taking a modified step.
#> Iteration 28 : loglikelihood = -9805.8338
#> Taking a modified step.
#> Iteration 29 : loglikelihood = -9805.8326
#> Taking a modified step.
#> Iteration 30 : loglikelihood = -9805.8316
#> Taking a modified step.
#> Iteration 31 : loglikelihood = -9805.8309
#> Taking a modified step.
#> Iteration 32 : loglikelihood = -9805.8304
#> Taking a modified step.
#> Iteration 33 : loglikelihood = -9805.83
#> Taking a modified step.
#> Iteration 34 : loglikelihood = -9805.8297
#> Taking a modified step.
#> Iteration 35 : loglikelihood = -9805.8295
#> Taking a modified step.
#> Iteration 36 : loglikelihood = -9805.8293
#> Taking a modified step.
#> Iteration 37 : loglikelihood = -9805.8291
#> Taking a modified step.
#> Iteration 38 : loglikelihood = -9805.829
#> Taking a modified step.
#> Iteration 39 : loglikelihood = -9805.8289
#> Taking a modified step.
#> Iteration 40 : loglikelihood = -9805.8289
#> Taking a modified step.
#> Iteration 41 : loglikelihood = -9805.8288
#> Taking a modified step.
#> Iteration 42 : loglikelihood = -9805.8288
#> Taking a modified step.
#> Iteration 43 : loglikelihood = -9805.8288
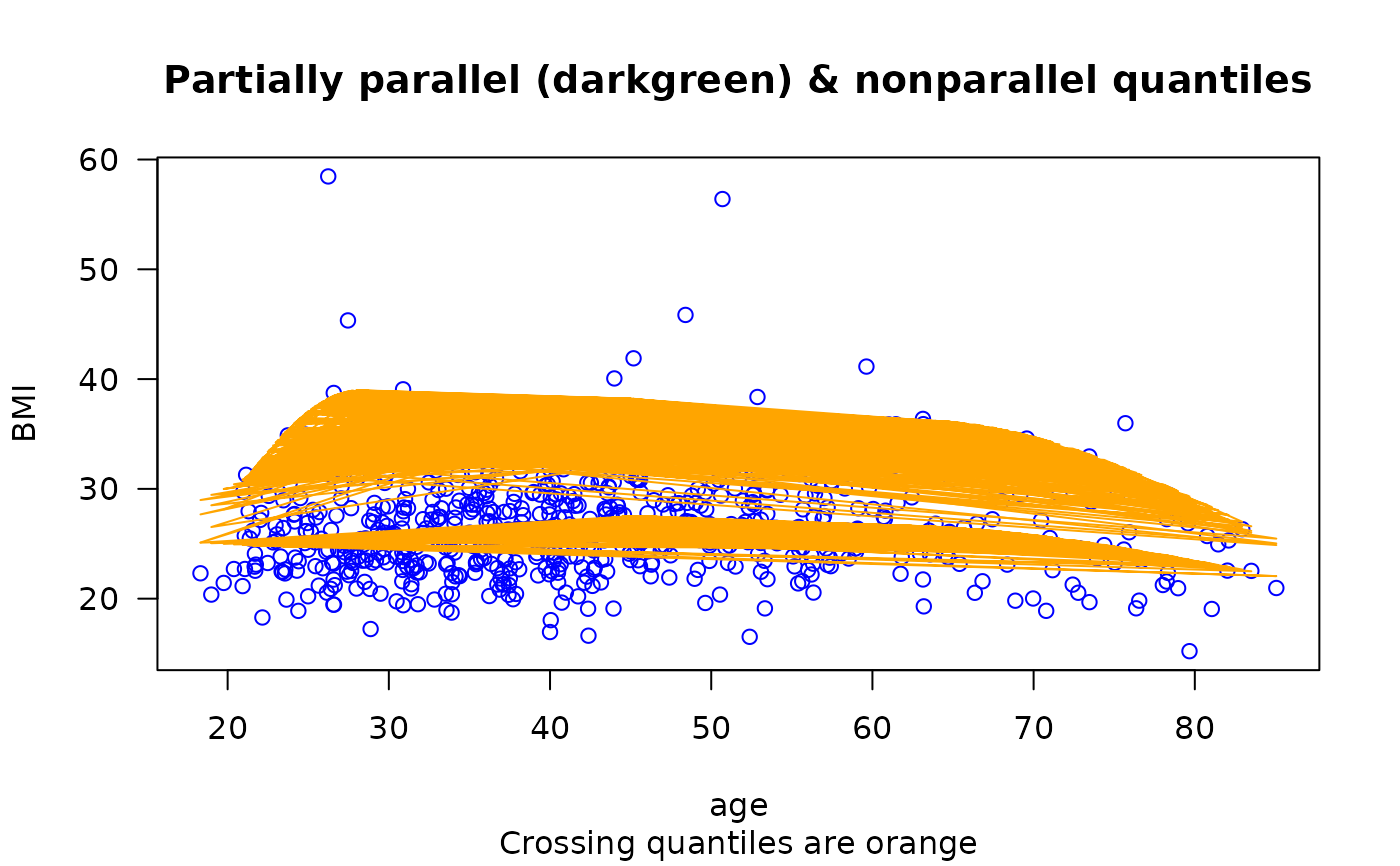
plot(BMI ~ age, bmi.nz, col = "blue", las = 1,
main = "Partially parallel (darkgreen) & nonparallel quantiles",
sub = "Crossing quantiles are orange")
is.crossing(fit1)
#> [1] TRUE
matlines(with(bmi.nz, age), fitted(fit1), lty = 1, col = "orange") # \dontrun{}
mytau <- c(50, 93, 95, 97) / 100 # Some quantiles are quite close
fit1 <- vglm(BMI ~ ns(age, 7), extlogF1(mytau), bmi.nz, trace = TRUE)
#> Taking a modified step.
#> Iteration 2 : loglikelihood = -9827.2672
#> Taking a modified step.
#> Iteration 3 : loglikelihood = -9821.8497
#> Taking a modified step.
#> Iteration 4 : loglikelihood = -9817.6926
#> Taking a modified step.
#> Iteration 5 : loglikelihood = -9814.6062
#> Taking a modified step.
#> Iteration 6 : loglikelihood = -9812.2572
#> Taking a modified step.
#> Iteration 7 : loglikelihood = -9810.5167
#> Taking a modified step.
#> Iteration 8 : loglikelihood = -9809.2165
#> Taking a modified step.
#> Iteration 9 : loglikelihood = -9808.2009
#> Taking a modified step.
#> Iteration 10 : loglikelihood = -9807.4706
#> Taking a modified step.
#> Iteration 11 : loglikelihood = -9807.0344
#> Taking a modified step.
#> Iteration 12 : loglikelihood = -9806.7858
#> Taking a modified step.
#> Iteration 13 : loglikelihood = -9806.6309
#> Taking a modified step.
#> Iteration 14 : loglikelihood = -9806.5045
#> Taking a modified step.
#> Iteration 15 : loglikelihood = -9806.3928
#> Taking a modified step.
#> Iteration 16 : loglikelihood = -9806.2922
#> Taking a modified step.
#> Iteration 17 : loglikelihood = -9806.2011
#> Taking a modified step.
#> Iteration 18 : loglikelihood = -9806.1193
#> Taking a modified step.
#> Iteration 19 : loglikelihood = -9806.047
#> Taking a modified step.
#> Iteration 20 : loglikelihood = -9805.9853
#> Taking a modified step.
#> Iteration 21 : loglikelihood = -9805.935
#> Taking a modified step.
#> Iteration 22 : loglikelihood = -9805.8964
#> Taking a modified step.
#> Iteration 23 : loglikelihood = -9805.8694
#> Taking a modified step.
#> Iteration 24 : loglikelihood = -9805.8525
#> Taking a modified step.
#> Iteration 25 : loglikelihood = -9805.8431
#> Taking a modified step.
#> Iteration 26 : loglikelihood = -9805.8382
#> Taking a modified step.
#> Iteration 27 : loglikelihood = -9805.8355
#> Taking a modified step.
#> Iteration 28 : loglikelihood = -9805.8338
#> Taking a modified step.
#> Iteration 29 : loglikelihood = -9805.8326
#> Taking a modified step.
#> Iteration 30 : loglikelihood = -9805.8316
#> Taking a modified step.
#> Iteration 31 : loglikelihood = -9805.8309
#> Taking a modified step.
#> Iteration 32 : loglikelihood = -9805.8304
#> Taking a modified step.
#> Iteration 33 : loglikelihood = -9805.83
#> Taking a modified step.
#> Iteration 34 : loglikelihood = -9805.8297
#> Taking a modified step.
#> Iteration 35 : loglikelihood = -9805.8295
#> Taking a modified step.
#> Iteration 36 : loglikelihood = -9805.8293
#> Taking a modified step.
#> Iteration 37 : loglikelihood = -9805.8291
#> Taking a modified step.
#> Iteration 38 : loglikelihood = -9805.829
#> Taking a modified step.
#> Iteration 39 : loglikelihood = -9805.8289
#> Taking a modified step.
#> Iteration 40 : loglikelihood = -9805.8289
#> Taking a modified step.
#> Iteration 41 : loglikelihood = -9805.8288
#> Taking a modified step.
#> Iteration 42 : loglikelihood = -9805.8288
#> Taking a modified step.
#> Iteration 43 : loglikelihood = -9805.8288
plot(BMI ~ age, bmi.nz, col = "blue", las = 1,
main = "Partially parallel (darkgreen) & nonparallel quantiles",
sub = "Crossing quantiles are orange")
is.crossing(fit1)
#> [1] TRUE
matlines(with(bmi.nz, age), fitted(fit1), lty = 1, col = "orange") # \dontrun{}