Mixture of Two Poisson Distributions
mix2poisson.RdEstimates the three parameters of a mixture of two Poisson distributions by maximum likelihood estimation.
mix2poisson(lphi = "logitlink", llambda = "loglink",
iphi = 0.5, il1 = NULL, il2 = NULL,
qmu = c(0.2, 0.8), nsimEIM = 100, zero = "phi")Arguments
- lphi, llambda
Link functions for the parameter \(\phi\) and \(\lambda\). See
Linksfor more choices.
- iphi
Initial value for \(\phi\), whose value must lie between 0 and 1.
- il1, il2
Optional initial value for \(\lambda_1\) and \(\lambda_2\). These values must be positive. The default is to compute initial values internally using the argument
qmu.- qmu
Vector with two values giving the probabilities relating to the sample quantiles for obtaining initial values for \(\lambda_1\) and \(\lambda_2\). The two values are fed in as the
probsargument intoquantile.- nsimEIM, zero
Details
The probability function can be loosely written as $$P(Y=y) = \phi \, Poisson(\lambda_1) + (1-\phi) \, Poisson(\lambda_2)$$ where \(\phi\) is the probability an observation belongs to the first group, and \(y=0,1,2,\ldots\). The parameter \(\phi\) satisfies \(0 < \phi < 1\). The mean of \(Y\) is \(\phi\lambda_1+(1-\phi)\lambda_2\) and this is returned as the fitted values. By default, the three linear/additive predictors are \((logit(\phi), \log(\lambda_1), \log(\lambda_2))^T\).
Value
An object of class "vglmff"
(see vglmff-class).
The object is used by modelling functions
such as vglm
and vgam.
Warning
This VGAM family function requires care for a successful
application.
In particular, good initial values are required because
of the presence of local solutions. Therefore running
this function with several different combinations of
arguments such as iphi, il1, il2,
qmu is highly recommended. Graphical methods such as
hist can be used as an aid.
With grouped data (i.e., using the weights argument)
one has to use a large value of nsimEIM;
see the example below.
This VGAM family function is experimental and should be used with care.
Note
The response must be integer-valued since
dpois is invoked.
Fitting this model successfully to data can be difficult
due to local solutions and ill-conditioned data. It pays to
fit the model several times with different initial values,
and check that the best fit looks reasonable. Plotting
the results is recommended. This function works better as
\(\lambda_1\) and \(\lambda_2\) become
more different. The default control argument trace =
TRUE is to encourage monitoring convergence.
See also
Examples
if (FALSE) # Example 1: simulated data
nn <- 1000
mu1 <- exp(2.5) # Also known as lambda1
mu2 <- exp(3)
(phi <- logitlink(-0.5, inverse = TRUE))
#> [1] 0.3775407
mdata <- data.frame(y = rpois(nn, ifelse(runif(nn) < phi, mu1, mu2)))
mfit <- vglm(y ~ 1, mix2poisson, data = mdata)
#> Iteration 1: loglikelihood = -1575.8839
#> Iteration 2: loglikelihood = -1575.6883
#> Iteration 3: loglikelihood = -1575.6868
#> Iteration 4: loglikelihood = -1575.6868
#> Iteration 5: loglikelihood = -1575.6868
coef(mfit, matrix = TRUE)
#> logitlink(phi) loglink(lambda1) loglink(lambda2)
#> (Intercept) -0.4095453 2.515517 3.01773
# Compare the results with the truth
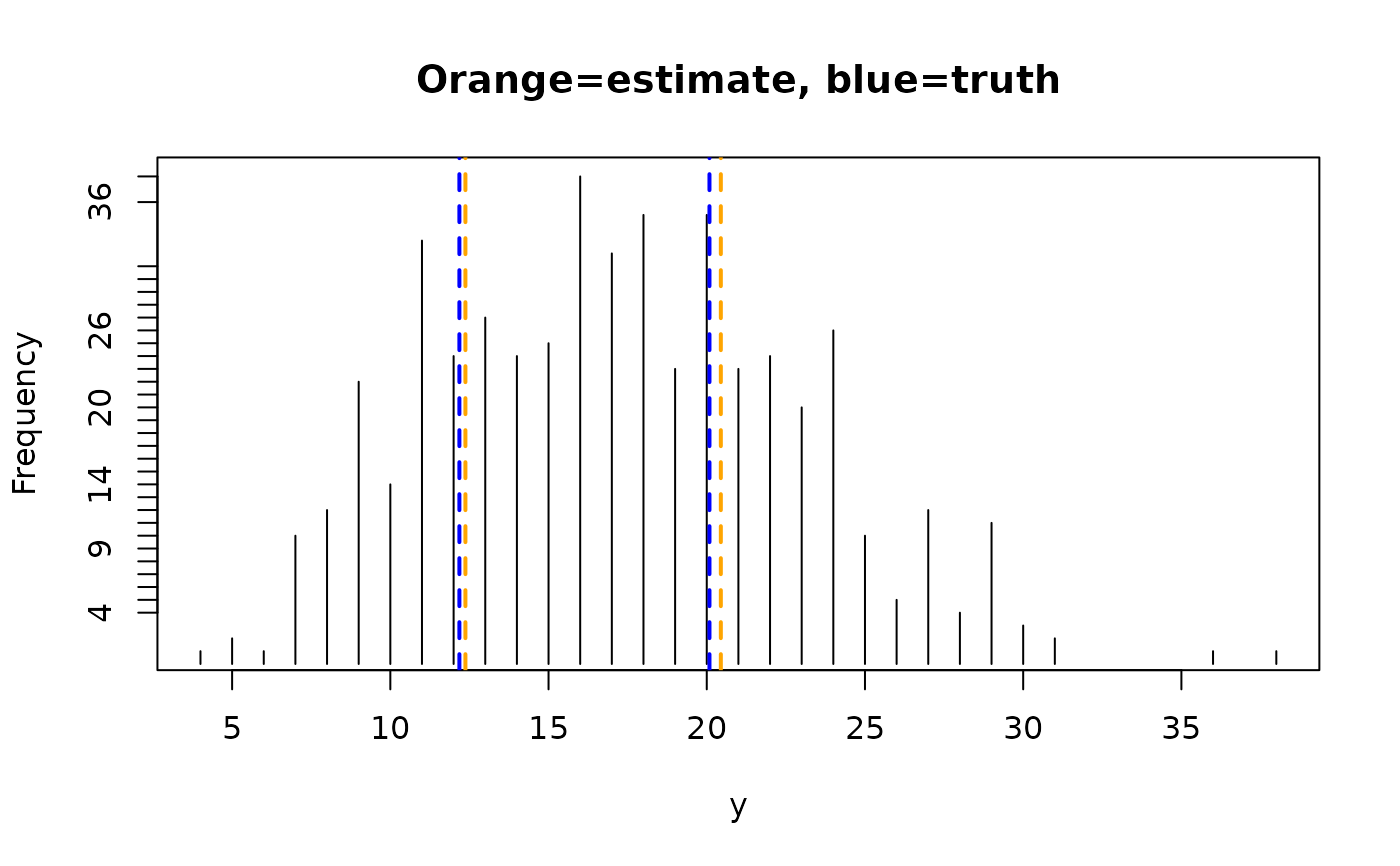
round(rbind('Estimated' = Coef(mfit), 'Truth' = c(phi, mu1, mu2)), 2)
#> phi lambda1 lambda2
#> Estimated 0.40 12.37 20.44
#> Truth 0.38 12.18 20.09
ty <- with(mdata, table(y))
plot(names(ty), ty, type = "h", main = "Orange=estimate, blue=truth",
ylab = "Frequency", xlab = "y")
abline(v = Coef(mfit)[-1], lty = 2, col = "orange", lwd = 2)
abline(v = c(mu1, mu2), lty = 2, col = "blue", lwd = 2)
 # Example 2: London Times data (Lange, 1997, p.31)
ltdata1 <- data.frame(deaths = 0:9,
freq = c(162,267,271, 185,111,61,27,8,3,1))
ltdata2 <- data.frame(y = with(ltdata1, rep(deaths, freq)))
# Usually this does not work well unless nsimEIM is large
Mfit <- vglm(deaths ~ 1, weight = freq, data = ltdata1,
mix2poisson(iphi=0.3, il1=1, il2=2.5, nsimEIM=5000))
#> Iteration 1: loglikelihood = -1989.9835
#> Applying Greenstadt modification to 10 matrices
#> Iteration 2: loglikelihood = -1993.7258
#> Taking a modified step..
#> Iteration 2 : loglikelihood = -1989.9756
#> Iteration 3: loglikelihood = -1989.9536
#> Iteration 4: loglikelihood = -1989.9575
#> Taking a modified step.
#> Iteration 4 : loglikelihood = -1989.9469
#> Iteration 5: loglikelihood = -1989.9459
#> Iteration 6: loglikelihood = -1989.9459
# This works better in general
Mfit = vglm(y ~ 1, mix2poisson(iphi=0.3, il1=1, il2=2.5), ltdata2)
#> Iteration 1: loglikelihood = -1990.00277
#> Iteration 2: loglikelihood = -1989.94587
#> Iteration 3: loglikelihood = -1989.94586
#> Iteration 4: loglikelihood = -1989.94586
coef(Mfit, matrix = TRUE)
#> logitlink(phi) loglink(lambda1) loglink(lambda2)
#> (Intercept) -0.5758701 0.2280049 0.9796043
Coef(Mfit)
#> phi lambda1 lambda2
#> 0.3598834 1.2560915 2.6634020
# \dontrun{}
# Example 2: London Times data (Lange, 1997, p.31)
ltdata1 <- data.frame(deaths = 0:9,
freq = c(162,267,271, 185,111,61,27,8,3,1))
ltdata2 <- data.frame(y = with(ltdata1, rep(deaths, freq)))
# Usually this does not work well unless nsimEIM is large
Mfit <- vglm(deaths ~ 1, weight = freq, data = ltdata1,
mix2poisson(iphi=0.3, il1=1, il2=2.5, nsimEIM=5000))
#> Iteration 1: loglikelihood = -1989.9835
#> Applying Greenstadt modification to 10 matrices
#> Iteration 2: loglikelihood = -1993.7258
#> Taking a modified step..
#> Iteration 2 : loglikelihood = -1989.9756
#> Iteration 3: loglikelihood = -1989.9536
#> Iteration 4: loglikelihood = -1989.9575
#> Taking a modified step.
#> Iteration 4 : loglikelihood = -1989.9469
#> Iteration 5: loglikelihood = -1989.9459
#> Iteration 6: loglikelihood = -1989.9459
# This works better in general
Mfit = vglm(y ~ 1, mix2poisson(iphi=0.3, il1=1, il2=2.5), ltdata2)
#> Iteration 1: loglikelihood = -1990.00277
#> Iteration 2: loglikelihood = -1989.94587
#> Iteration 3: loglikelihood = -1989.94586
#> Iteration 4: loglikelihood = -1989.94586
coef(Mfit, matrix = TRUE)
#> logitlink(phi) loglink(lambda1) loglink(lambda2)
#> (Intercept) -0.5758701 0.2280049 0.9796043
Coef(Mfit)
#> phi lambda1 lambda2
#> 0.3598834 1.2560915 2.6634020
# \dontrun{}