The CCl4 Inhalation Model
ccl4model.RdThe CCl4 inhalation model implemented in .Fortran
ccl4model(times, y, parms, ...)Arguments
- times
time sequence for which the model has to be integrated.
- y
the initial values for the state variables ("AI", "AAM", "AT", "AF", "AL", "CLT" and "AM"), in that order.
- parms
vector or list holding the ccl4 model parameters; see the example for the order in which these have to be defined.
- ...
any other parameters passed to the integrator
ode(which solves the model).
Details
The model is implemented primarily to demonstrate the linking of FORTRAN with R-code.
The source can be found in the /doc/examples/dynload subdirectory of the
package.
See also
Examples
## =================
## Parameter values
## =================
Pm <- c(
## Physiological parameters
BW = 0.182, # Body weight (kg)
QP = 4.0 , # Alveolar ventilation rate (hr^-1)
QC = 4.0 , # Cardiac output (hr^-1)
VFC = 0.08, # Fraction fat tissue (kg/(kg/BW))
VLC = 0.04, # Fraction liver tissue (kg/(kg/BW))
VMC = 0.74, # Fraction of muscle tissue (kg/(kg/BW))
QFC = 0.05, # Fractional blood flow to fat ((hr^-1)/QC
QLC = 0.15, # Fractional blood flow to liver ((hr^-1)/QC)
QMC = 0.32, # Fractional blood flow to muscle ((hr^-1)/QC)
## Chemical specific parameters for chemical
PLA = 16.17, # Liver/air partition coefficient
PFA = 281.48, # Fat/air partition coefficient
PMA = 13.3, # Muscle/air partition coefficient
PTA = 16.17, # Viscera/air partition coefficient
PB = 5.487, # Blood/air partition coefficient
MW = 153.8, # Molecular weight (g/mol)
VMAX = 0.04321671, # Max. velocity of metabolism (mg/hr) -calibrated
KM = 0.4027255, # Michaelis-Menten constant (mg/l) -calibrated
## Parameters for simulated experiment
CONC = 1000, # Inhaled concentration
KL = 0.02, # Loss rate from empty chamber /hr
RATS = 1.0, # Number of rats enclosed in chamber
VCHC = 3.8 # Volume of closed chamber (l)
)
## ================
## State variables
## ================
y <- c(
AI = 21, # total mass , mg
AAM = 0,
AT = 0,
AF = 0,
AL = 0,
CLT = 0, # area under the conc.-time curve in the liver
AM = 0 # the amount metabolized (AM)
)
## ==================
## Model application
## ==================
times <- seq(0, 6, by = 0.1)
## initial inhaled concentration-calibrated
conc <- c(26.496, 90.197, 245.15, 951.46)
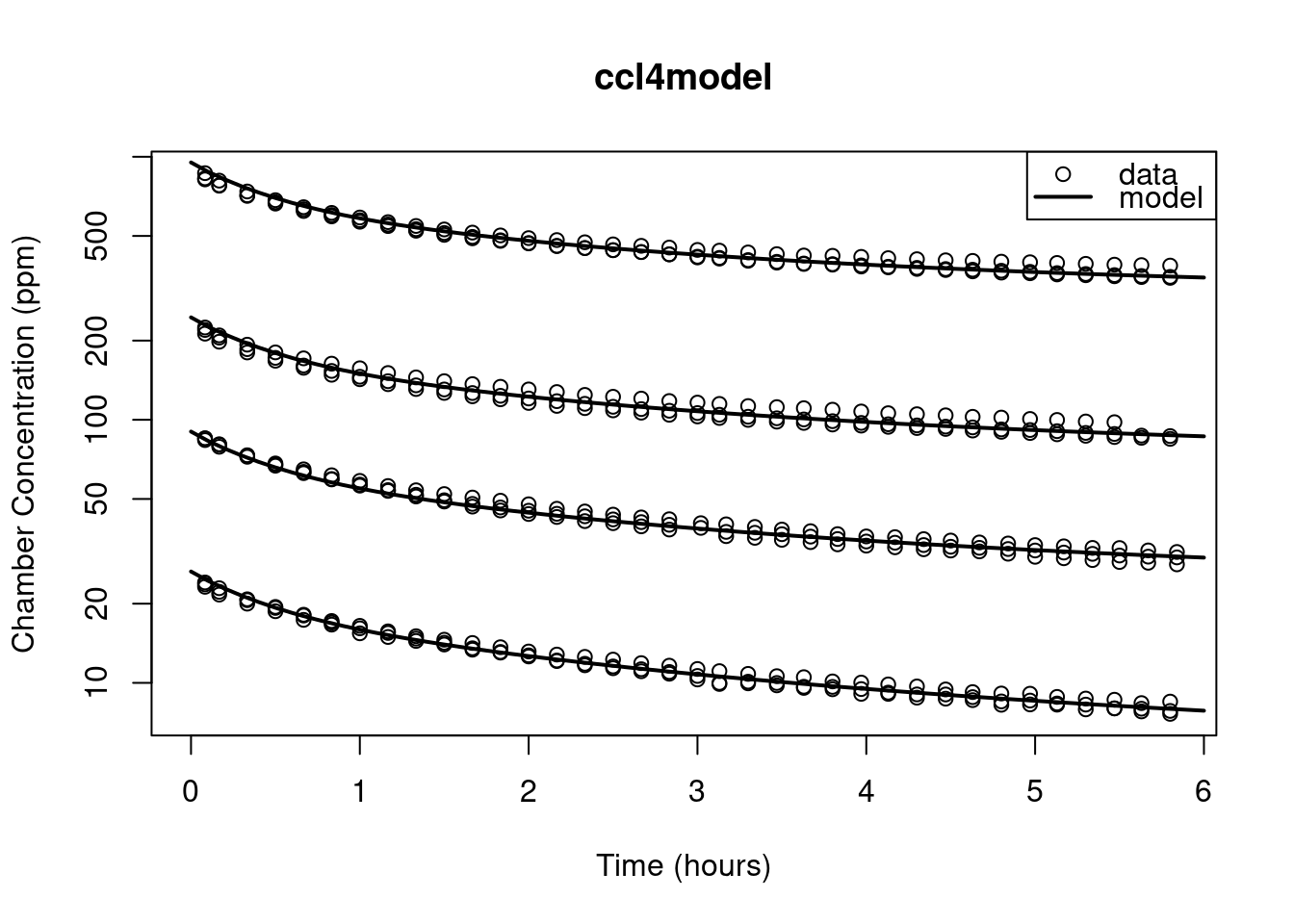
plot(ChamberConc ~ time, data = ccl4data, xlab = "Time (hours)",
xlim = range(c(0, ccl4data$time)),
ylab = "Chamber Concentration (ppm)",
log = "y", main = "ccl4model")
for (cc in conc) {
Pm["CONC"] <- cc
VCH <- Pm[["VCHC"]] - Pm[["RATS"]] * Pm[["BW"]]
AI0 <- VCH * Pm[["CONC"]] * Pm[["MW"]]/24450
y["AI"] <- AI0
## run the model:
out <- as.data.frame(ccl4model(times, y, Pm))
lines(out$time, out$CP, lwd = 2)
}
legend("topright", lty = c(NA, 1), pch = c(1, NA), lwd = c(NA, 2),
legend = c("data", "model"))
 ## ==================================
## An example with tracer injection
## ==================================
## every day, a conc of 2 is added to AI.
## 1. implemented as a data.frame
eventdat <- data.frame(var = rep("AI", 6), time = 1:6 ,
value = rep(1, 6), method = rep("add", 6))
eventdat
#> var time value method
#> 1 AI 1 1 add
#> 2 AI 2 1 add
#> 3 AI 3 1 add
#> 4 AI 4 1 add
#> 5 AI 5 1 add
#> 6 AI 6 1 add
print(system.time(
out <-ccl4model(times, y, Pm, events = list(data = eventdat))
))
#> user system elapsed
#> 0.003 0.000 0.003
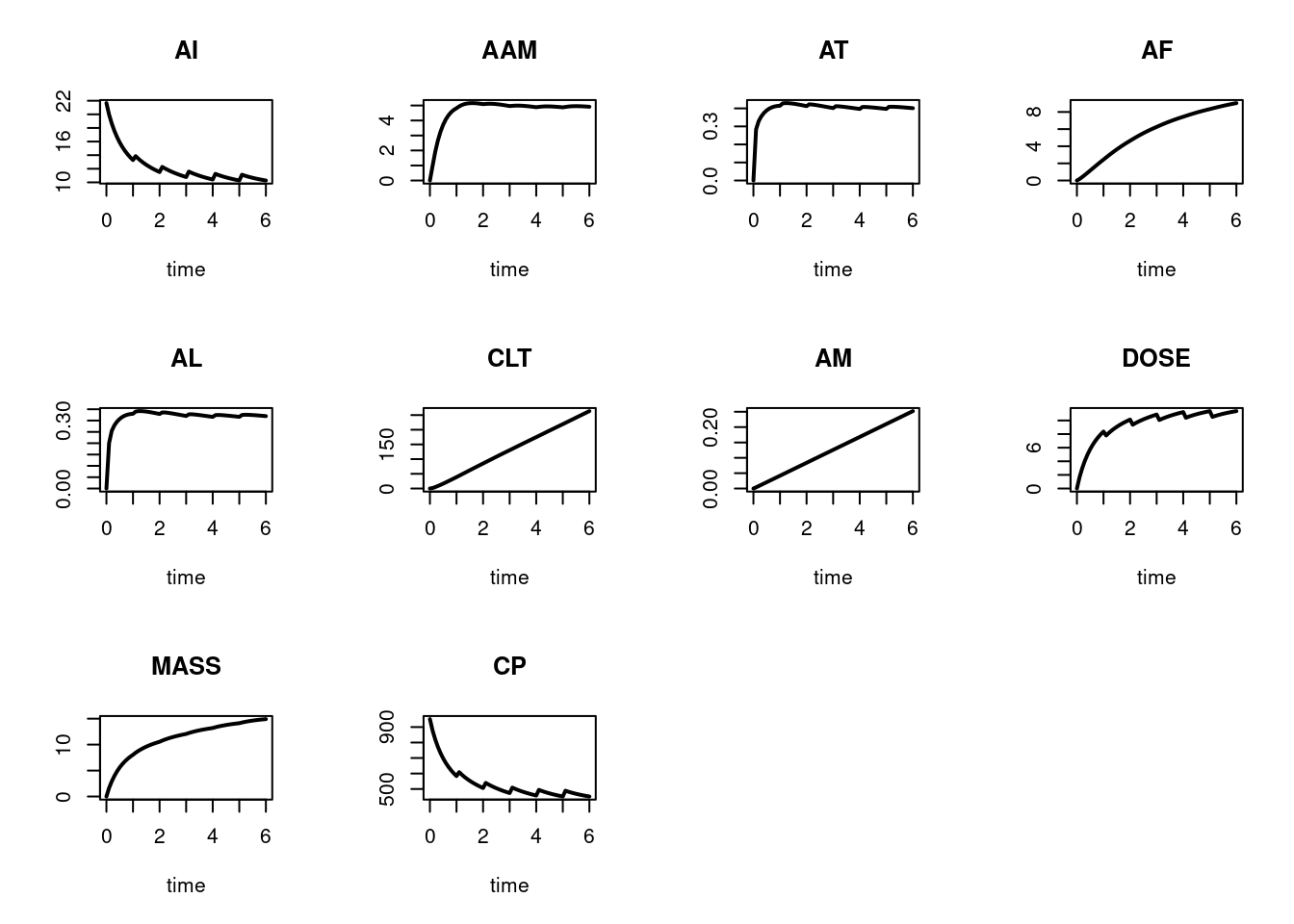
plot(out, mfrow = c(3, 4), type = "l", lwd = 2)
# 2. implemented as a function in a DLL!
print(system.time(
out2 <-ccl4model(times, y, Pm, events = list(func = "eventfun", time = 1:6))
))
#> user system elapsed
#> 0.001 0.000 0.001
plot(out2, mfrow=c(3, 4), type = "l", lwd = 2)
## ==================================
## An example with tracer injection
## ==================================
## every day, a conc of 2 is added to AI.
## 1. implemented as a data.frame
eventdat <- data.frame(var = rep("AI", 6), time = 1:6 ,
value = rep(1, 6), method = rep("add", 6))
eventdat
#> var time value method
#> 1 AI 1 1 add
#> 2 AI 2 1 add
#> 3 AI 3 1 add
#> 4 AI 4 1 add
#> 5 AI 5 1 add
#> 6 AI 6 1 add
print(system.time(
out <-ccl4model(times, y, Pm, events = list(data = eventdat))
))
#> user system elapsed
#> 0.003 0.000 0.003
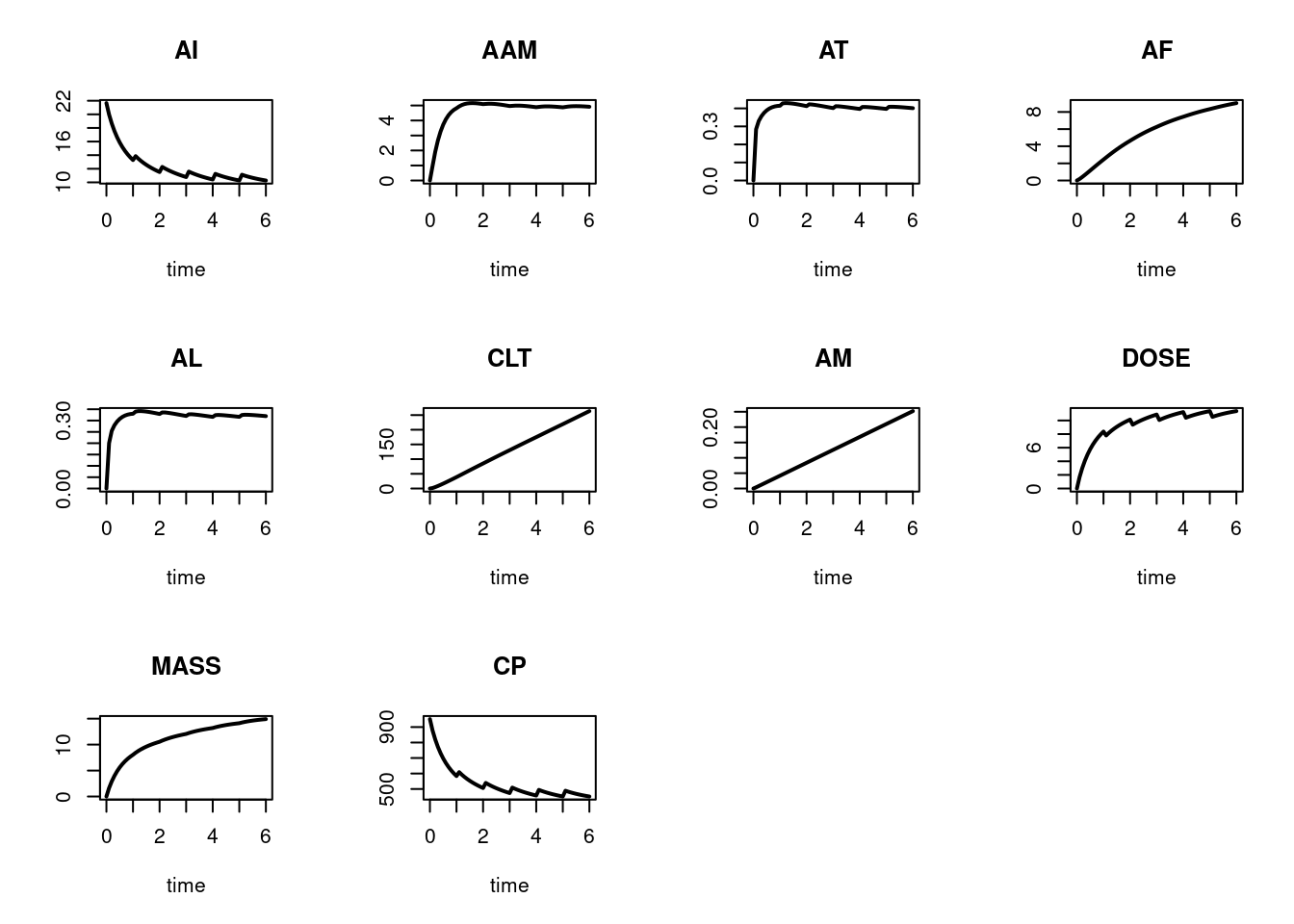
plot(out, mfrow = c(3, 4), type = "l", lwd = 2)
# 2. implemented as a function in a DLL!
print(system.time(
out2 <-ccl4model(times, y, Pm, events = list(func = "eventfun", time = 1:6))
))
#> user system elapsed
#> 0.001 0.000 0.001
plot(out2, mfrow=c(3, 4), type = "l", lwd = 2)