Kullback-Leibler Divergence
KLdiv.RdEstimate the Kullback-Leibler divergence of several distributions.
# S4 method for class 'matrix'
KLdiv(object, eps = 10^-4, overlap = TRUE,...)
# S4 method for class 'flexmix'
KLdiv(object, method = c("continuous", "discrete"), ...)Arguments
- object
See Methods section below.
- method
The method to be used; "continuous" determines the Kullback-Leibler divergence between the unweighted theoretical component distributions and the unweighted posterior probabilities at the observed points are used by "discrete".
- eps
Probabilities below this threshold are replaced by this threshold for numerical stability.
- overlap
Logical, do not determine the KL divergence for those pairs where for each point at least one of the densities has a value smaller than
eps.- ...
Passed to the matrix method.
Methods
- object = "matrix":
Takes as input a matrix of density values with one row per observation and one column per distribution.
- object = "flexmix":
Returns the Kullback-Leibler divergence of the mixture components.
Details
Estimates $$\int f(x) (\log f(x) - \log g(x)) dx$$ for distributions with densities \(f()\) and \(g()\).
Value
A matrix of KL divergences where the rows correspond to using the respective distribution as \(f()\) in the formula above.
Note
The density functions are modified to have equal support.
A weight of at least eps is given to each
observation point for the modified densities.
References
S. Kullback and R. A. Leibler. On information and sufficiency.The Annals of Mathematical Statistics, 22(1), 79–86, 1951.
Friedrich Leisch. Exploring the structure of mixture model components. In Jaromir Antoch, editor, Compstat 2004–Proceedings in Computational Statistics, 1405–1412. Physika Verlag, Heidelberg, Germany, 2004. ISBN 3-7908-1554-3.
Examples
## Gaussian and Student t are much closer to each other than
## to the uniform:
x <- seq(-3, 3, length = 200)
y <- cbind(u = dunif(x), n = dnorm(x), t = dt(x, df = 10))
matplot(x, y, type = "l")
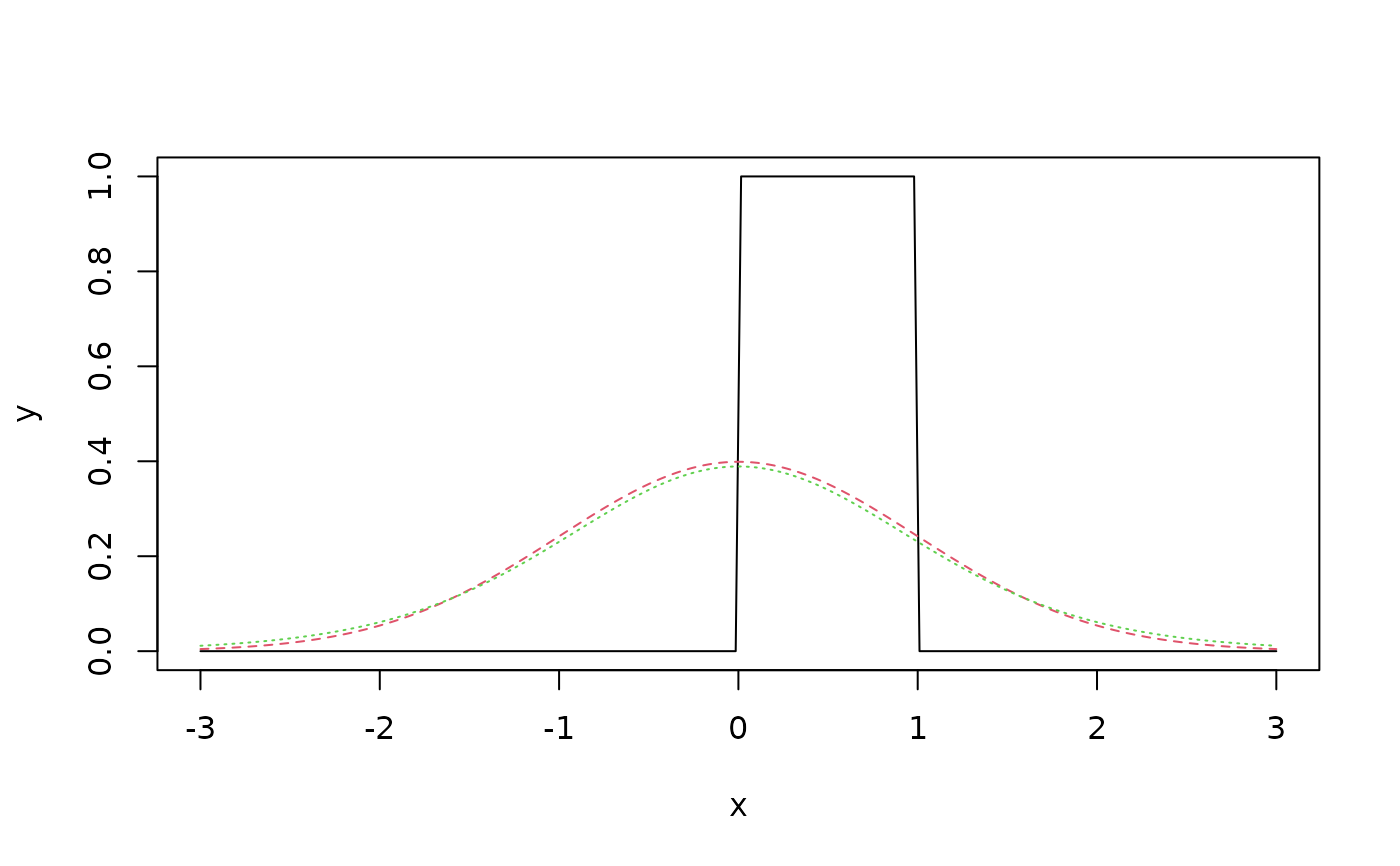 KLdiv(y)
#> u n t
#> u 0.000000 1.082016066 1.107696457
#> n 4.661417 0.000000000 0.004314432
#> t 4.685997 0.005138101 0.000000000
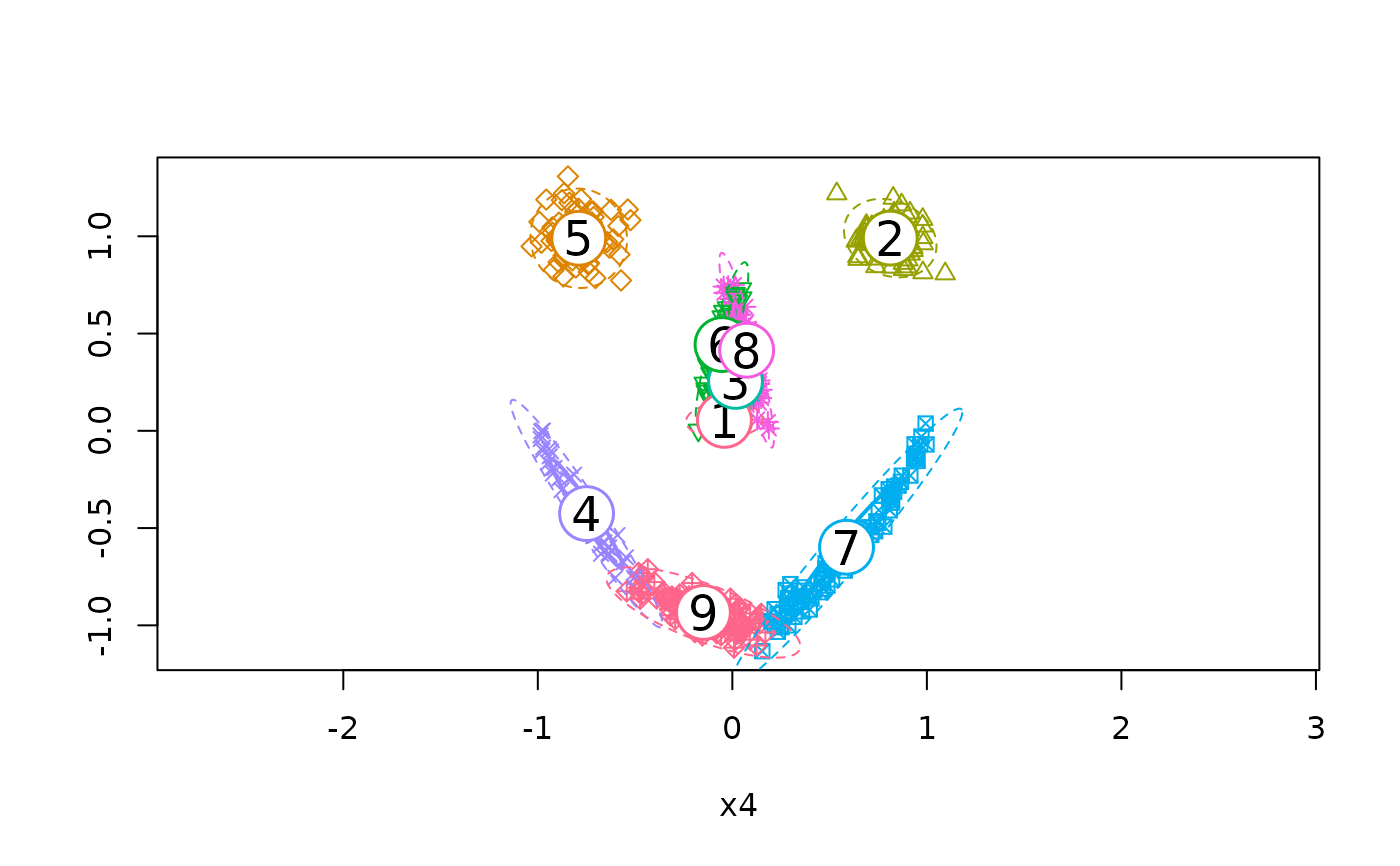
if (require("mlbench")) {
set.seed(2606)
x <- mlbench.smiley()$x
model1 <- flexmix(x ~ 1, k = 9, model = FLXmclust(diag = FALSE),
control = list(minprior = 0))
plotEll(model1, x)
KLdiv(model1)
}
#> Loading required package: mlbench
KLdiv(y)
#> u n t
#> u 0.000000 1.082016066 1.107696457
#> n 4.661417 0.000000000 0.004314432
#> t 4.685997 0.005138101 0.000000000
if (require("mlbench")) {
set.seed(2606)
x <- mlbench.smiley()$x
model1 <- flexmix(x ~ 1, k = 9, model = FLXmclust(diag = FALSE),
control = list(minprior = 0))
plotEll(model1, x)
KLdiv(model1)
}
#> Loading required package: mlbench
 #> [,1] [,2] [,3] [,4] [,5] [,6] [,7]
#> [1,] 0.00000 127.38549 10.465786 347.39410 65.96873 15.68134 238.37734
#> [2,] 355.09493 0.00000 515.453871 2046.99689 124.57769 342.86906 216.96622
#> [3,] 16.37309 91.53808 0.000000 485.00219 55.37133 11.91903 285.47909
#> [4,] 135.20050 346.13459 473.194050 0.00000 94.69614 180.33986 401.47033
#> [5,] 374.62909 141.08019 245.535712 274.52425 0.00000 524.48359 1259.40178
#> [6,] 63.83343 77.94779 7.378457 538.34468 40.15853 0.00000 394.59000
#> [7,] 222.54714 214.59542 170.061156 555.34119 206.81012 597.40484 0.00000
#> [8,] 60.69815 68.54197 7.659784 625.11702 51.71476 19.63367 326.94140
#> [9,] 352.43050 381.10288 206.550561 34.20101 187.22028 115.71277 49.44981
#> [,8] [,9]
#> [1,] 44.405475 131.53573
#> [2,] 701.249827 645.13681
#> [3,] 9.320418 194.35845
#> [4,] 965.009194 19.41957
#> [5,] 464.799392 365.19535
#> [6,] 20.048972 251.81501
#> [7,] 156.737792 66.82421
#> [8,] 0.000000 257.36165
#> [9,] 332.601354 0.00000
#> [,1] [,2] [,3] [,4] [,5] [,6] [,7]
#> [1,] 0.00000 127.38549 10.465786 347.39410 65.96873 15.68134 238.37734
#> [2,] 355.09493 0.00000 515.453871 2046.99689 124.57769 342.86906 216.96622
#> [3,] 16.37309 91.53808 0.000000 485.00219 55.37133 11.91903 285.47909
#> [4,] 135.20050 346.13459 473.194050 0.00000 94.69614 180.33986 401.47033
#> [5,] 374.62909 141.08019 245.535712 274.52425 0.00000 524.48359 1259.40178
#> [6,] 63.83343 77.94779 7.378457 538.34468 40.15853 0.00000 394.59000
#> [7,] 222.54714 214.59542 170.061156 555.34119 206.81012 597.40484 0.00000
#> [8,] 60.69815 68.54197 7.659784 625.11702 51.71476 19.63367 326.94140
#> [9,] 352.43050 381.10288 206.550561 34.20101 187.22028 115.71277 49.44981
#> [,8] [,9]
#> [1,] 44.405475 131.53573
#> [2,] 701.249827 645.13681
#> [3,] 9.320418 194.35845
#> [4,] 965.009194 19.41957
#> [5,] 464.799392 365.19535
#> [6,] 20.048972 251.81501
#> [7,] 156.737792 66.82421
#> [8,] 0.000000 257.36165
#> [9,] 332.601354 0.00000