Point and interval summaries for tidy data frames of draws from distributions
Source:R/point_interval.R
point_interval.RdTranslates draws from distributions in a (possibly grouped) data frame into point and interval summaries (or set of point and interval summaries, if there are multiple groups in a grouped data frame).
Supports automatic partial function application.
point_interval(
.data,
...,
.width = 0.95,
.point = median,
.interval = qi,
.simple_names = TRUE,
na.rm = FALSE,
.exclude = c(".chain", ".iteration", ".draw", ".row"),
.prob
)
# Default S3 method
point_interval(
.data,
...,
.width = 0.95,
.point = median,
.interval = qi,
.simple_names = TRUE,
na.rm = FALSE,
.exclude = c(".chain", ".iteration", ".draw", ".row"),
.prob
)
# S3 method for class 'tbl_df'
point_interval(.data, ...)
# S3 method for class 'numeric'
point_interval(
.data,
...,
.width = 0.95,
.point = median,
.interval = qi,
.simple_names = FALSE,
na.rm = FALSE,
.exclude = c(".chain", ".iteration", ".draw", ".row"),
.prob
)
# S3 method for class 'rvar'
point_interval(
.data,
...,
.width = 0.95,
.point = median,
.interval = qi,
.simple_names = TRUE,
na.rm = FALSE
)
# S3 method for class 'distribution'
point_interval(
.data,
...,
.width = 0.95,
.point = median,
.interval = qi,
.simple_names = TRUE,
na.rm = FALSE
)
qi(x, .width = 0.95, .prob, na.rm = FALSE)
ll(x, .width = 0.95, na.rm = FALSE)
ul(x, .width = 0.95, na.rm = FALSE)
hdi(
x,
.width = 0.95,
na.rm = FALSE,
...,
density = density_bounded(trim = TRUE),
n = 4096,
.prob
)
Mode(x, na.rm = FALSE, ...)
# Default S3 method
Mode(
x,
na.rm = FALSE,
...,
density = density_bounded(trim = TRUE),
n = 2001,
weights = NULL
)
# S3 method for class 'rvar'
Mode(x, na.rm = FALSE, ...)
# S3 method for class 'distribution'
Mode(x, na.rm = FALSE, ...)
hdci(x, .width = 0.95, na.rm = FALSE)
mean_qi(.data, ..., .width = 0.95)
median_qi(.data, ..., .width = 0.95)
mode_qi(.data, ..., .width = 0.95)
mean_ll(.data, ..., .width = 0.95)
median_ll(.data, ..., .width = 0.95)
mode_ll(.data, ..., .width = 0.95)
mean_ul(.data, ..., .width = 0.95)
median_ul(.data, ..., .width = 0.95)
mode_ul(.data, ..., .width = 0.95)
mean_hdi(.data, ..., .width = 0.95)
median_hdi(.data, ..., .width = 0.95)
mode_hdi(.data, ..., .width = 0.95)
mean_hdci(.data, ..., .width = 0.95)
median_hdci(.data, ..., .width = 0.95)
mode_hdci(.data, ..., .width = 0.95)Arguments
- .data
<data.frame | grouped_df> Data frame (or grouped data frame as returned by
dplyr::group_by()) that contains draws to summarize.- ...
<bare language> Column names or expressions that, when evaluated in the context of
.data, represent draws to summarize. If this is empty, then by default all columns that are not group columns and which are not in.exclude(by default".chain",".iteration",".draw", and".row") will be summarized. These columns can be numeric, distributional objects,posterior::rvars, or list columns of numeric values to summarise.- .width
<numeric> vector of probabilities to use that determine the widths of the resulting intervals. If multiple probabilities are provided, multiple rows per group are generated, each with a different probability interval (and value of the corresponding
.widthcolumn).- .point
<function> Point summary function, which takes a vector and returns a single value, e.g.
mean,median, orMode.- .interval
<function> Interval function, which takes a vector and a probability (
.width) and returns a two-element vector representing the lower and upper bound of an interval; e.g.qi,hdi- .simple_names
<scalar logical> When
TRUEand only a single column / vector is to be summarized, use the name.lowerfor the lower end of the interval and.upperfor the upper end. If.datais a vector and this isTRUE, this will also set the column name of the point summary to.value. WhenFALSEand.datais a data frame, names the lower and upper intervals for each columnxx.lowerandx.upper. WhenFALSEand.datais a vector, uses the naming schemey,yminandymax(for use with ggplot).- na.rm
<scalar logical> Should
NAvalues be stripped before the computation proceeds? IfFALSE(the default), any vectors to be summarized that containNAwill result in point and interval summaries equal toNA.- .exclude
<character> Vector of names of columns to be excluded from summarization if no column names are specified to be summarized in
.... Default ignores several meta-data column names used in ggdist and tidybayes.- .prob
Deprecated. Use
.widthinstead.- x
<numeric> Vector to summarize (for interval functions:
qi(),hdi(), etc)- density
<function | string> For
hdi()andMode(), the kernel density estimator to use, either as a function (e.g.density_bounded,density_unbounded) or as a string giving the suffix to a function that starts withdensity_(e.g."bounded"or"unbounded"). The default,"bounded", uses the bounded density estimator ofdensity_bounded(), which itself estimates the bounds of the distribution, and tends to work well on both bounded and unbounded data.- n
<scalar numeric> For
hdi()andMode(), the number of points to use to estimate highest-density intervals or modes.- weights
<numeric | NULL> For
Mode(), an optional vector, which (if notNULL) is of the same length asxand provides weights for each element ofx.
Value
A data frame containing point summaries and intervals, with at least one column corresponding
to the point summary, one to the lower end of the interval, one to the upper end of the interval, the
width of the interval (.width), the type of point summary (.point), and the type of interval (.interval).
Details
If .data is a data frame, then ... is a list of bare names of
columns (or expressions derived from columns) of .data, on which
the point and interval summaries are derived. Column expressions are processed
using the tidy evaluation framework (see rlang::eval_tidy()).
For a column named x, the resulting data frame will have a column
named x containing its point summary. If there is a single
column to be summarized and .simple_names is TRUE, the output will
also contain columns .lower (the lower end of the interval),
.upper (the upper end of the interval).
Otherwise, for every summarized column x, the output will contain
x.lower (the lower end of the interval) and x.upper (the upper
end of the interval). Finally, the output will have a .width column
containing the' probability for the interval on each output row.
If .data includes groups (see e.g. dplyr::group_by()),
the points and intervals are calculated within the groups.
If .data is a vector, ... is ignored and the result is a
data frame with one row per value of .width and three columns:
y (the point summary), ymin (the lower end of the interval),
ymax (the upper end of the interval), and .width, the probability
corresponding to the interval. This behavior allows point_interval
and its derived functions (like median_qi, mean_qi, mode_hdi, etc)
to be easily used to plot intervals in ggplot stats using methods like
stat_eye(), stat_halfeye(), or stat_summary().
median_qi, mode_hdi, etc are short forms for
point_interval(..., .point = median, .interval = qi), etc.
qi yields the quantile interval (also known as the percentile interval or
equi-tailed interval) as a 1x2 matrix.
hdi yields the highest-density interval(s) (also known as the highest posterior
density interval). Note: If the distribution is multimodal, hdi may return multiple
intervals for each probability level (these will be spread over rows). You may wish to use
hdci (below) instead if you want a single highest-density interval, with the caveat that when
the distribution is multimodal hdci is not a highest-density interval.
hdci yields the highest-density continuous interval, also known as the shortest
probability interval. Note: If the distribution is multimodal, this may not actually
be the highest-density interval (there may be a higher-density
discontinuous interval, which can be found using hdi).
ll and ul yield lower limits and upper limits, respectively (where the opposite
limit is set to either Inf or -Inf).
Examples
library(dplyr)
library(ggplot2)
set.seed(123)
rnorm(1000) %>%
median_qi()
#> y ymin ymax .width .point .interval
#> 1 0.009209639 -1.941554 2.037887 0.95 median qi
data.frame(x = rnorm(1000)) %>%
median_qi(x, .width = c(.50, .80, .95))
#> # A tibble: 3 × 6
#> x .lower .upper .width .point .interval
#> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
#> 1 0.0549 -0.653 0.753 0.5 median qi
#> 2 0.0549 -1.24 1.34 0.8 median qi
#> 3 0.0549 -1.99 1.91 0.95 median qi
data.frame(
x = rnorm(1000),
y = rnorm(1000, mean = 2, sd = 2)
) %>%
median_qi(x, y)
#> x x.lower x.upper y y.lower y.upper .width .point
#> 1 -0.05057431 -2.012529 1.934141 1.983618 -1.946229 5.947635 0.95 median
#> .interval
#> 1 qi
data.frame(
x = rnorm(1000),
group = "a"
) %>%
rbind(data.frame(
x = rnorm(1000, mean = 2, sd = 2),
group = "b")
) %>%
group_by(group) %>%
median_qi(.width = c(.50, .80, .95))
#> # A tibble: 6 × 7
#> group x .lower .upper .width .point .interval
#> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
#> 1 a -0.0328 -0.707 0.636 0.5 median qi
#> 2 b 2.06 0.759 3.44 0.5 median qi
#> 3 a -0.0328 -1.27 1.23 0.8 median qi
#> 4 b 2.06 -0.559 4.48 0.8 median qi
#> 5 a -0.0328 -2.00 1.84 0.95 median qi
#> 6 b 2.06 -1.75 5.91 0.95 median qi
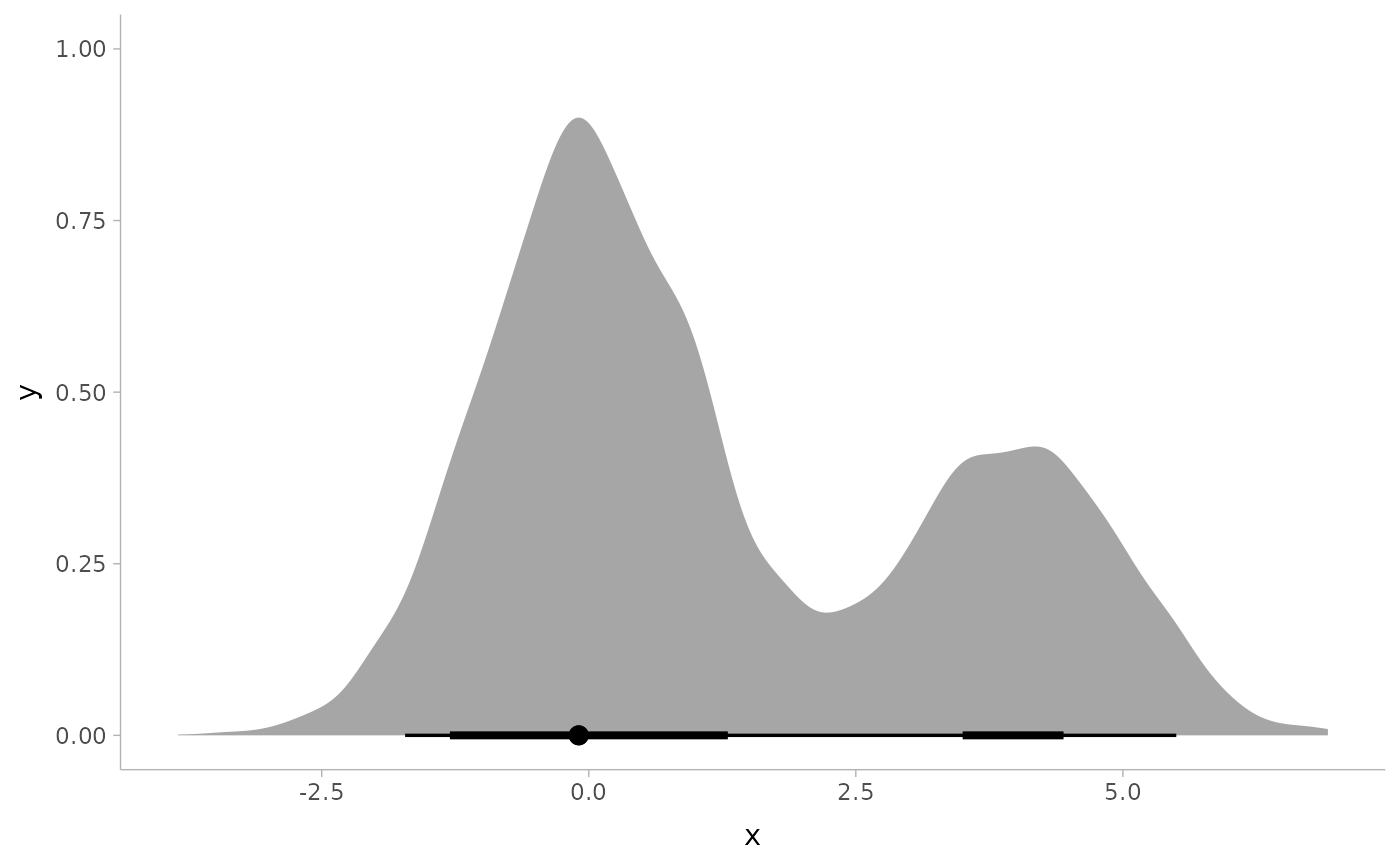
multimodal_draws = data.frame(
x = c(rnorm(5000, 0, 1), rnorm(2500, 4, 1))
)
multimodal_draws %>%
mode_hdi(.width = c(.66, .95))
#> # A tibble: 3 × 6
#> x .lower .upper .width .point .interval
#> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
#> 1 -0.0938 -1.30 1.30 0.66 mode hdi
#> 2 -0.0938 3.50 4.44 0.66 mode hdi
#> 3 -0.0938 -1.72 5.50 0.95 mode hdi
multimodal_draws %>%
ggplot(aes(x = x, y = 0)) +
stat_halfeye(point_interval = mode_hdi, .width = c(.66, .95))