Calculate and plot smoothed time series.
panel.tskernel.RdPlot time series smoothed by discrete symmetric smoothing kernels. These kernels can be used to smooth time series objects. Options include moving averages, triangular filters, or approximately Gaussian filters.
panel.tskernel(x, y, ...,
width = NROW(x) %/% 10 + 1, n = 300,
c = 1, sides = 2, circular = FALSE,
kern = kernel("daniell",
rep(floor((width/sides) / sqrt(c)), c)))
simpleSmoothTs(x, ...)
# Default S3 method
simpleSmoothTs(x, ...,
width = NROW(x) %/% 10 + 1, n = NROW(x),
c = 1, sides = 2, circular = FALSE,
kern = kernel("daniell",
rep(floor((width/sides)/sqrt(c)), c)))
# S3 method for class 'zoo'
simpleSmoothTs(x, ..., n = NROW(x))Arguments
- x, y
data points. Should define a regular, ordered series. A time series object can be passed as the first argument, in which case
ycan be omitted. Thexargument given tosimpleSmoothTsis allowed to be a multivariate time series, i.e. to have multiple columns.- ...
further arguments passed on to
panel.lines.- width
nominal width of the smoothing kernel in time steps. In the default case, which is a simple moving average, this is the actual width. When
c > 1the number of time steps used in the kernel increases but the equivalent bandwidth stays the same. If only past values are used (withsides = 1) thenwidthrefers to one side of the symmetric kernel.- n
approximate number of time steps desired for the result. If this is less than the length of
x, the smoothed time series will be aggregated by averaging blocks of (an integer number of) time steps, and this aggregated series will be centered with respect to the original series.- c
smoothness of the kernel:
c = 1is a moving average,c = 2is a triangular kernel,c = 3and higher approximate smooth Gaussian kernels.cis actually the number of times to recursively convolve a simple moving average kernel with itself. The kernel size is adjusted to maintain a constant equivalent bandwidth ascincreases.- sides
if
sides=1the smoothed series is calculed from past values only (using one half of the symmetric kernel); ifsides=2it is centred around lag 0.- circular
to treat the data as circular (periodic).
- kern
a
tskernelobject; if given, this over-rideswidthandc.
Note
The author is not an expert on time series theory.
Examples
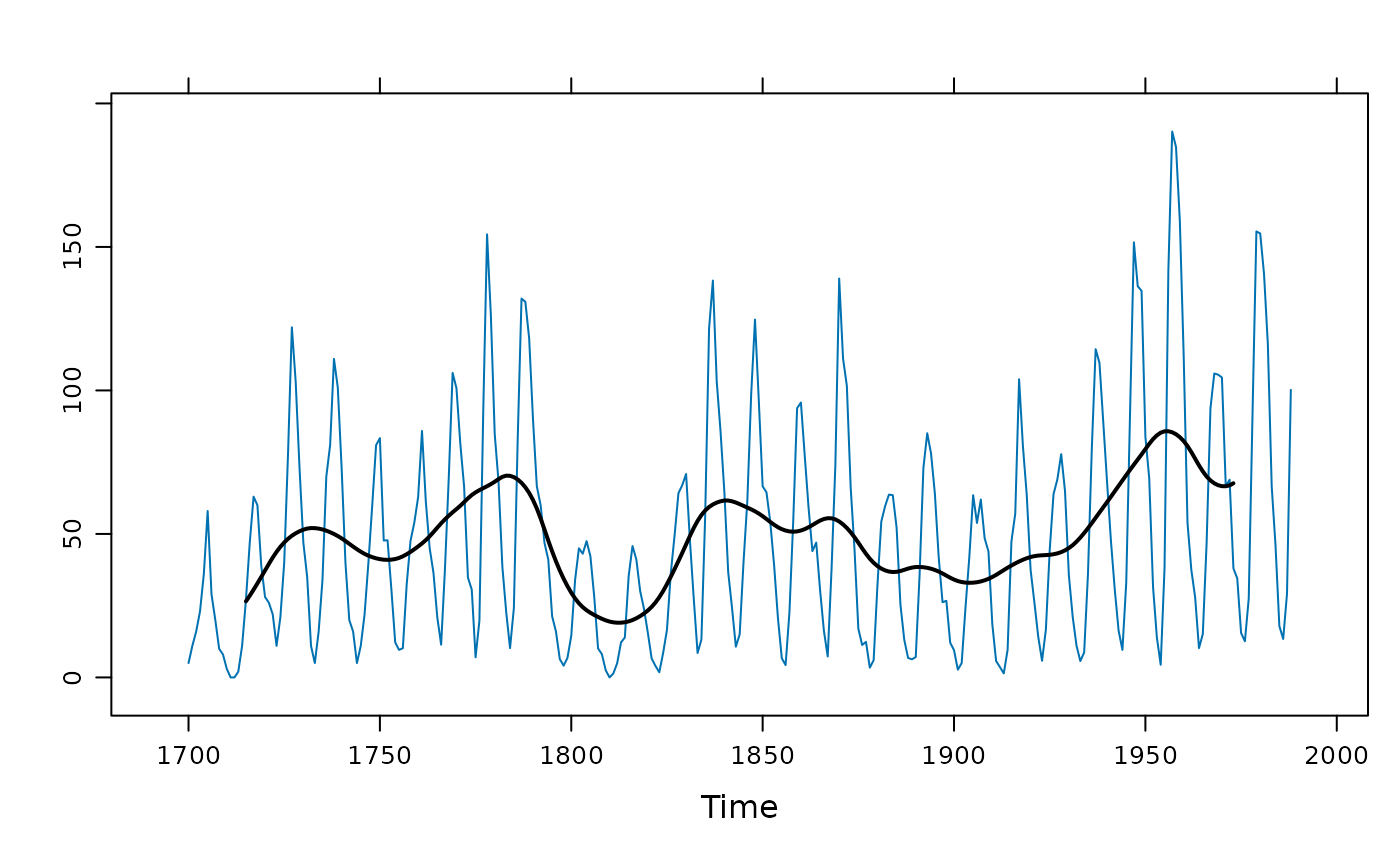
## a Gaussian-like filter (contrast with c = 1 or c = 2)
xyplot(sunspot.year) +
layer(panel.tskernel(x, y, width = 20, c = 3, col = 1, lwd = 2))
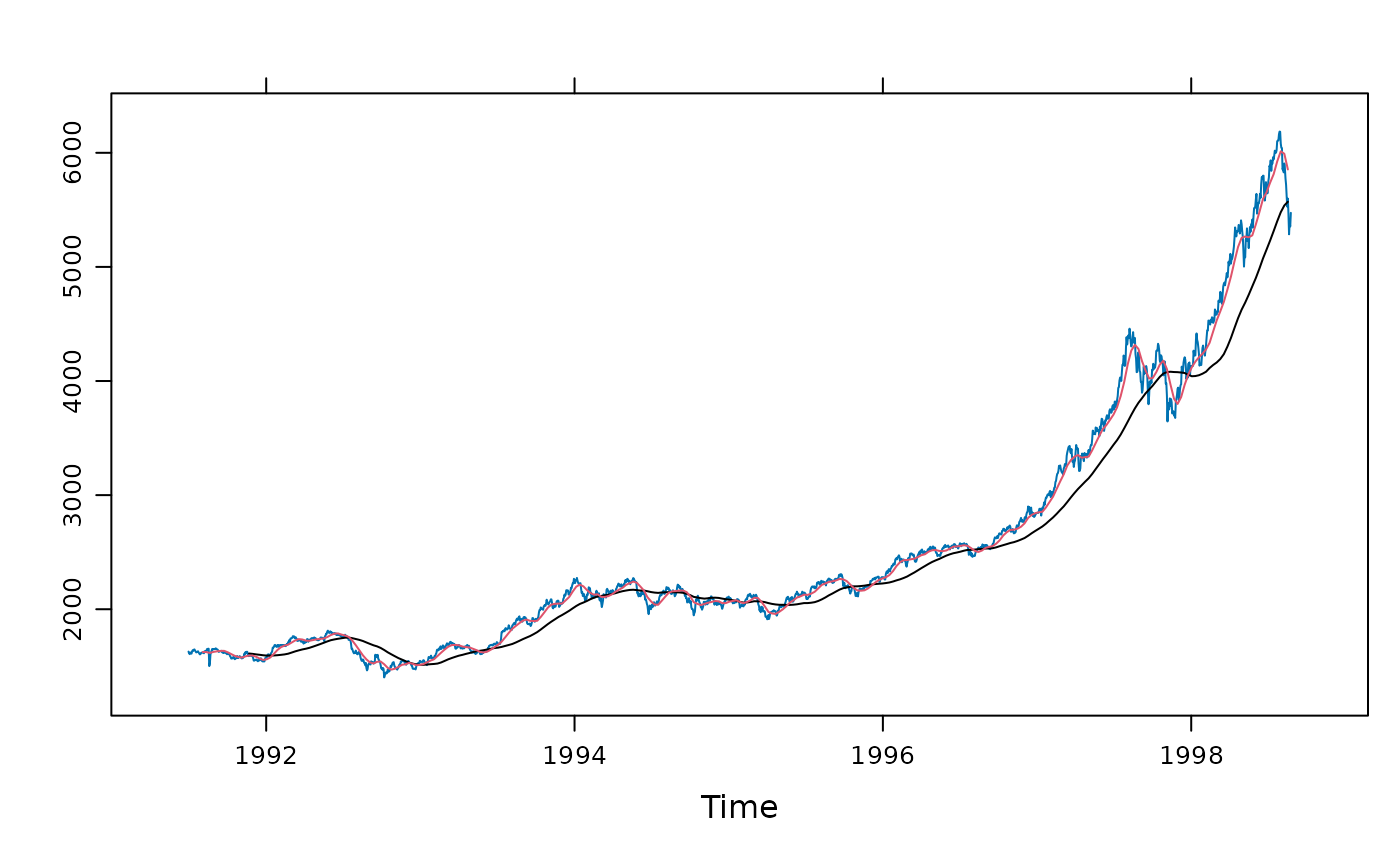
 ## example from ?kernel:
## long and short moving averages, backwards in time
xyplot(EuStockMarkets[,1]) +
layer(panel.tskernel(x, y, width = 100, col = 1, sides = 1)) +
layer(panel.tskernel(x, y, width = 20, col = 2, sides = 1))
## example from ?kernel:
## long and short moving averages, backwards in time
xyplot(EuStockMarkets[,1]) +
layer(panel.tskernel(x, y, width = 100, col = 1, sides = 1)) +
layer(panel.tskernel(x, y, width = 20, col = 2, sides = 1))
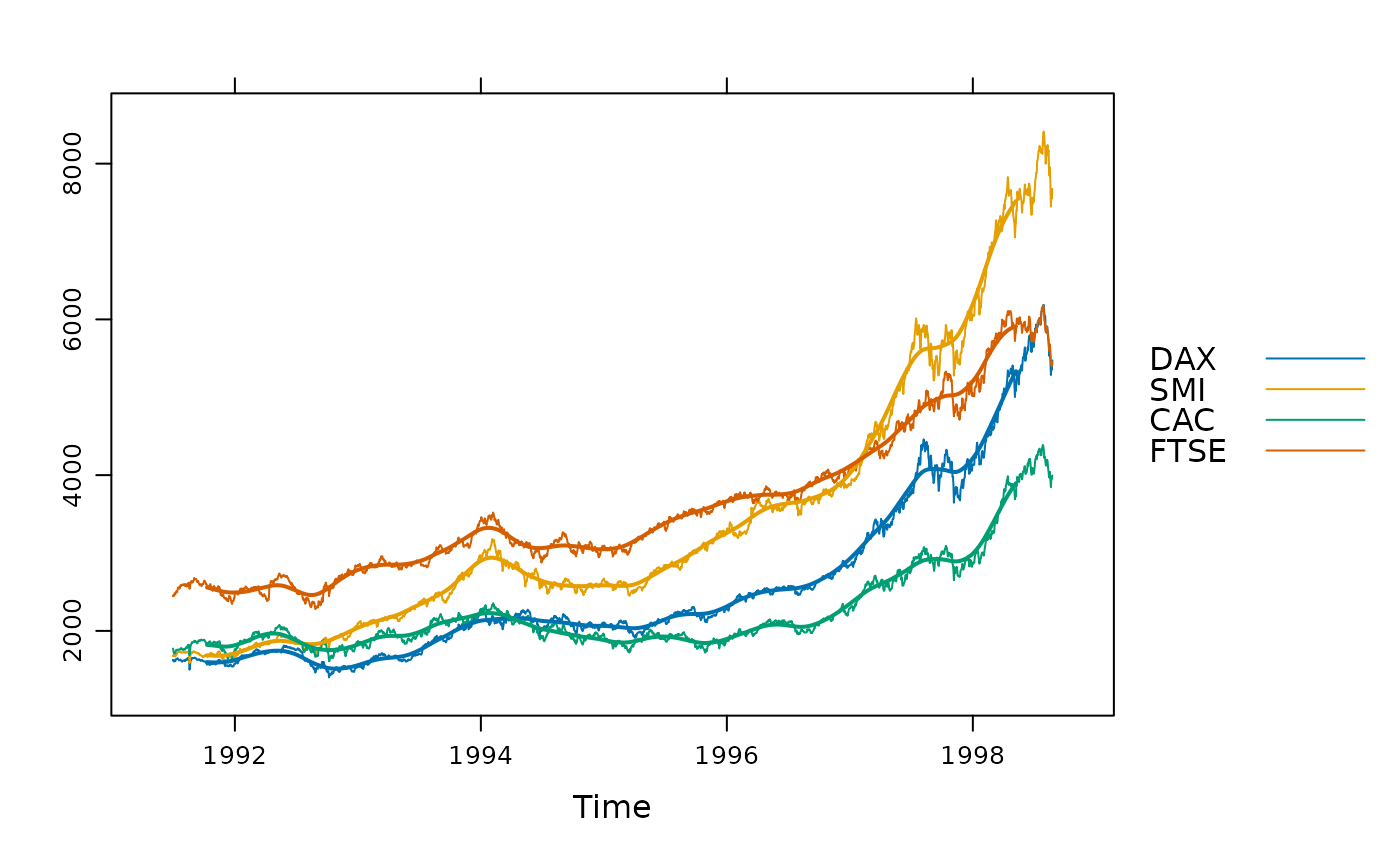
 ## per group, with a triangular filter
xyplot(EuStockMarkets, superpose = TRUE) +
glayer(panel.tskernel(..., width = 100, c = 2),
theme = simpleTheme(lwd = 2))
## per group, with a triangular filter
xyplot(EuStockMarkets, superpose = TRUE) +
glayer(panel.tskernel(..., width = 100, c = 2),
theme = simpleTheme(lwd = 2))
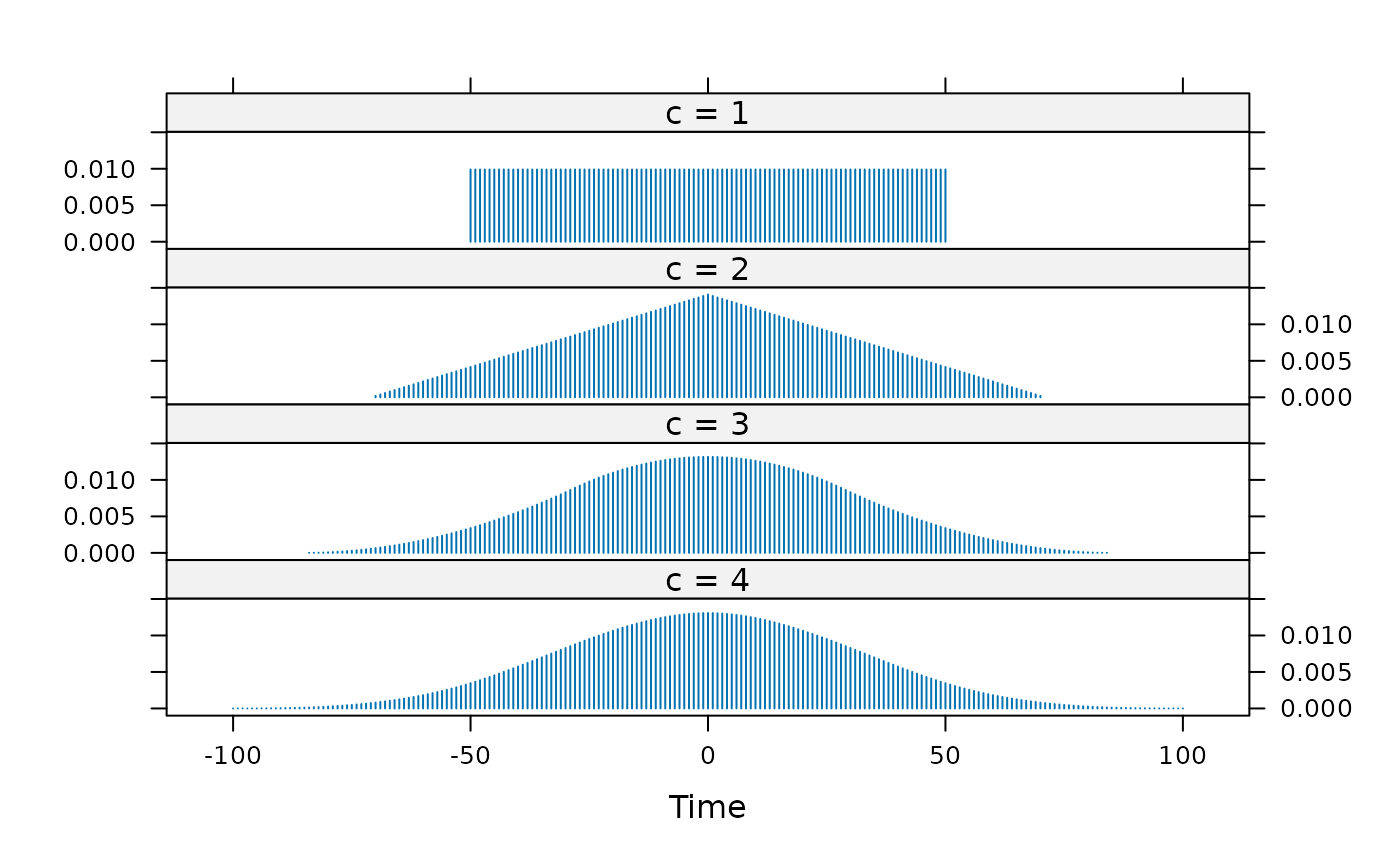
 ## plot the actual kernels used; note adjustment of width
width = 100
kdat <- lapply(1:4, function(c) {
k <- kernel("daniell", rep(floor(0.5*width / sqrt(c)), c))
## demonstrate that the effective bandwidth stays the same:
message("c = ", c, ": effective bandwidth = ", bandwidth.kernel(k))
## represent the kernel as a time series, for plotting
ts(k[-k$m:k$m], start = -k$m)
})
#> c = 1: effective bandwidth = 29.1561885940761
#> c = 2: effective bandwidth = 28.9841910933069
#> c = 3: effective bandwidth = 28.4970758733828
#> c = 4: effective bandwidth = 29.440618200031
names(kdat) <- paste("c =", 1:4)
xyplot(do.call(ts.union, kdat), type = "h",
scales = list(y = list(relation = "same")))
## plot the actual kernels used; note adjustment of width
width = 100
kdat <- lapply(1:4, function(c) {
k <- kernel("daniell", rep(floor(0.5*width / sqrt(c)), c))
## demonstrate that the effective bandwidth stays the same:
message("c = ", c, ": effective bandwidth = ", bandwidth.kernel(k))
## represent the kernel as a time series, for plotting
ts(k[-k$m:k$m], start = -k$m)
})
#> c = 1: effective bandwidth = 29.1561885940761
#> c = 2: effective bandwidth = 28.9841910933069
#> c = 3: effective bandwidth = 28.4970758733828
#> c = 4: effective bandwidth = 29.440618200031
names(kdat) <- paste("c =", 1:4)
xyplot(do.call(ts.union, kdat), type = "h",
scales = list(y = list(relation = "same")))