01 - Introduction to ‘pbkrtest’
Søren Højsgaard and Ulrich Halekoh
a01-pbkrtest.rmd## Loading required package: pbkrtest## Loading required package: lme4## Loading required package: MatrixPackage version: 0.5.4
Introduction
The data is a list of two vectors, giving the wear of shoes of materials A and B for one foot each of ten boys.
data(shoes, package="MASS")
shoes## $A
## [1] 13.2 8.2 10.9 14.3 10.7 6.6 9.5 10.8 8.8 13.3
##
## $B
## [1] 14.0 8.8 11.2 14.2 11.8 6.4 9.8 11.3 9.3 13.6A plot reveals that boys wear their shoes differently.
plot(A ~ 1, data=shoes, col="red",lwd=2, pch=1, ylab="wear", xlab="boy")
points(B ~ 1, data=shoes, col="blue", lwd=2, pch=2)
points(I((A + B) / 2) ~ 1, data=shoes, pch="-", lwd=2)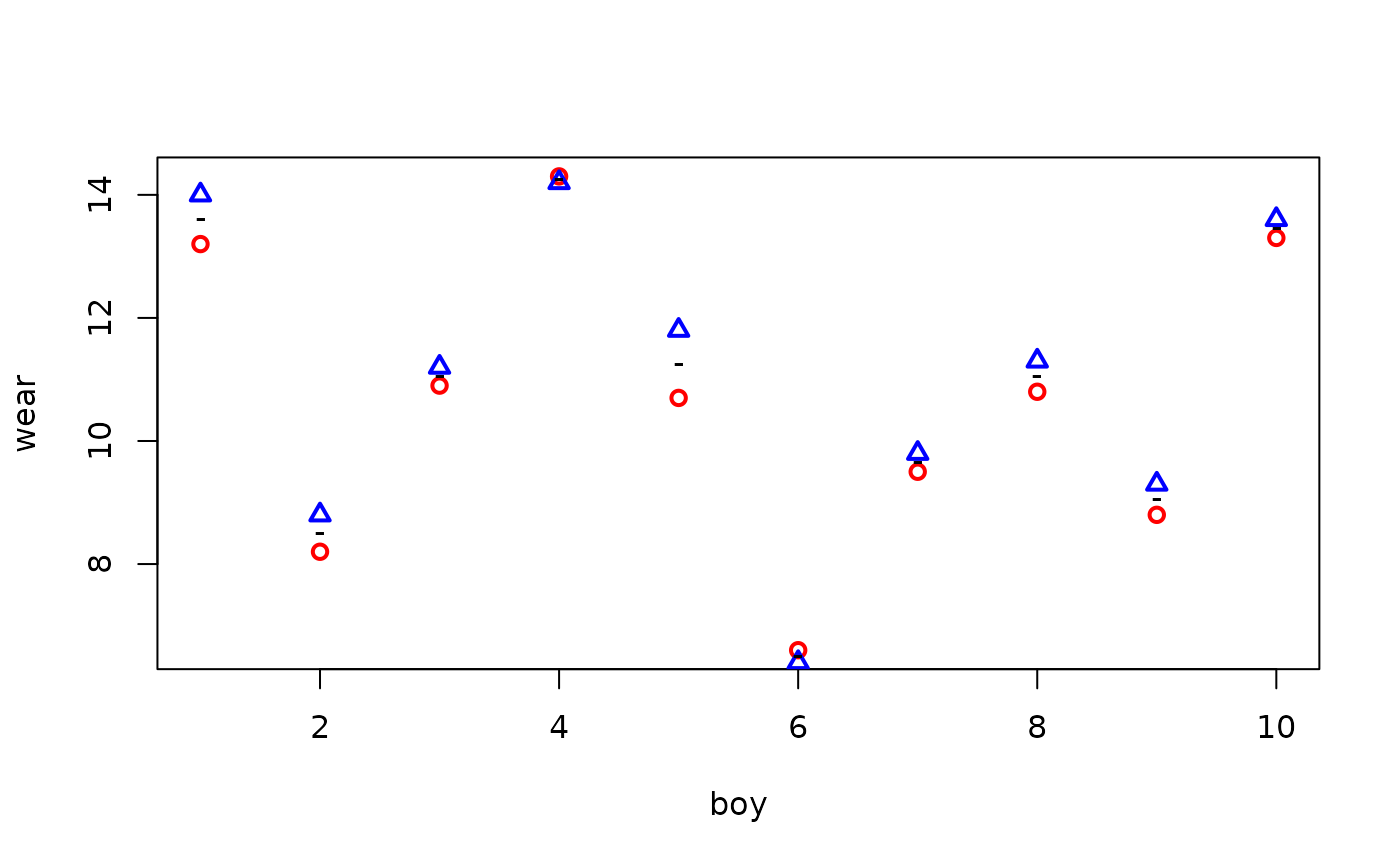
One option for testing the effect of materials is to make a paired –test, e.g. as:
## # A tibble: 1 × 8
## estimate statistic p.value parameter conf.low conf.high method alternative
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 -0.41 -3.35 0.00854 9 -0.687 -0.133 Paired t-… two.sidedTo work with data in a mixed model setting we create a dataframe, and for later use we also create an imbalanced version of data:
boy <- rep(1:10, 2)
boyf<- factor(letters[boy])
material <- factor(c(rep("A", 10), rep("B", 10)))
## Balanced data:
shoe.bal <- data.frame(wear=unlist(shoes), boy=boy, boyf=boyf, material=material)
head(shoe.bal)## wear boy boyf material
## A1 13.2 1 a A
## A2 8.2 2 b A
## A3 10.9 3 c A
## A4 14.3 4 d A
## A5 10.7 5 e A
## A6 6.6 6 f A
## Imbalanced data; delete (boy=1, material=1) and (boy=2, material=b)
shoe.imbal <- shoe.bal[-c(1, 12),]We fit models to the two datasets:
lmm1.bal <- lmer( wear ~ material + (1|boyf), data=shoe.bal)
lmm0.bal <- update(lmm1.bal, .~. - material)
lmm1.imbal <- lmer(wear ~ material + (1|boyf), data=shoe.imbal)
lmm0.imbal <- update(lmm1.imbal, .~. - material)The asymptotic likelihood ratio test shows stronger significance than the –test:
## refitting model(s) with ML (instead of REML)## # A tibble: 2 × 9
## term npar AIC BIC logLik minus2logL statistic df p.value
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 lmm0.bal 3 67.9 70.9 -31.0 61.9 NA NA NA
## 2 lmm1.bal 4 61.8 65.8 -26.9 53.8 8.09 1 0.00445## refitting model(s) with ML (instead of REML)## # A tibble: 2 × 9
## term npar AIC BIC logLik minus2logL statistic df p.value
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 lmm0.imbal 3 63.9 66.5 -28.9 57.9 NA NA NA
## 2 lmm1.imbal 4 60.8 64.3 -26.4 52.8 5.09 1 0.0240Kenward–Roger approach
The Kenward–Roger approximation is exact in certain balanced designs in the sense that the approximation produces the same result as the paired –test.
## # A tibble: 1 × 5
## term stat df ddf p.value
## <chr> <dbl> <int> <dbl> <dbl>
## 1 KR_Ftest 11.2 1 9.00 0.00854## # A tibble: 3 × 5
## term stat df ddf p.value
## <chr> <dbl> <int> <dbl> <dbl>
## 1 LRT 8.12 1 NA 0.00438
## 2 KR_Ftest 11.2 1 9.00 0.00854
## 3 KR_FtestU 11.2 1 9.00 0.00854For the imbalanced data we get
## # A tibble: 1 × 5
## term stat df ddf p.value
## <chr> <dbl> <int> <dbl> <dbl>
## 1 KR_Ftest 5.99 1 7.02 0.0442## # A tibble: 3 × 5
## term stat df ddf p.value
## <chr> <dbl> <int> <dbl> <dbl>
## 1 LRT 5.12 1 NA 0.0237
## 2 KR_Ftest 5.99 1 7.02 0.0442
## 3 KR_FtestU 5.99 1 7.02 0.0442Notice that the Kenward-Roger approximation gives results similar to but not identical to the paired –test when the two boys are removed:
## # A tibble: 1 × 8
## estimate statistic p.value parameter conf.low conf.high method alternative
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 -0.337 -2.39 0.0483 7 -0.672 -0.00328 Paired t-… two.sidedSatterthwaite approach
The Satterthwaite approximation is exact in certain balanced designs in the sense that the approximation produces the same result as the paired –test.
sat.bal <- SATmodcomp(lmm1.bal, lmm0.bal)
sat.bal |> tidy()## # A tibble: 1 × 5
## term stat df ddf p.value
## <chr> <dbl> <int> <dbl> <dbl>
## 1 SAT_Ftest 11.2 1 9.00 0.00854
sat.imbal <- SATmodcomp(lmm1.imbal, lmm0.imbal)
sat.imbal |> tidy()## # A tibble: 1 × 5
## term stat df ddf p.value
## <chr> <dbl> <int> <dbl> <dbl>
## 1 SAT_Ftest 6.00 1 7.01 0.0441Parametric bootstrap
Parametric bootstrap provides an alternative but many simulations are often needed to provide credible results (also many more than shown here; in this connection it can be useful to exploit that computations can be made en parallel, see the documentation):
## # A tibble: 1 × 5
## term stat df ddf p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 PBtest 8.12 NA NA 0.00399## # A tibble: 3 × 5
## term stat df ddf p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 LRT 8.12 1 NA 0.00438
## 2 PBtest 8.12 NA NA 0.00399
## 3 NA NA NA NA NAFor the imbalanced data, the result is similar to the result from the paired –test.
## # A tibble: 1 × 5
## term stat df ddf p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 PBtest 5.12 NA NA 0.0659## # A tibble: 3 × 5
## term stat df ddf p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 LRT 5.12 1 NA 0.0237
## 2 PBtest 5.12 NA NA 0.0659
## 3 NA NA NA NA NAMatrices for random effects
The matrices involved in the random effects can be obtained with
shoe3 <- subset(shoe.bal, boy<=5)
shoe3 <- shoe3[order(shoe3$boy), ]
lmm1 <- lmer( wear ~ material + (1|boyf), data=shoe3 )
str( SG <- get_SigmaG( lmm1 ), max=2)## List of 3
## $ Sigma :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
## $ G :List of 2
## ..$ :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
## ..$ :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
## $ n.ggamma: int 2
round( SG$Sigma*10 )## 10 x 10 sparse Matrix of class "dgCMatrix"## [[ suppressing 10 column names 'A1', 'B1', 'A2' ... ]]##
## A1 53 52 . . . . . . . .
## B1 52 53 . . . . . . . .
## A2 . . 53 52 . . . . . .
## B2 . . 52 53 . . . . . .
## A3 . . . . 53 52 . . . .
## B3 . . . . 52 53 . . . .
## A4 . . . . . . 53 52 . .
## B4 . . . . . . 52 53 . .
## A5 . . . . . . . . 53 52
## B5 . . . . . . . . 52 53
SG$G## [[1]]
## 10 x 10 sparse Matrix of class "dgCMatrix"## [[ suppressing 10 column names 'A1', 'B1', 'A2' ... ]]##
## A1 1 1 . . . . . . . .
## B1 1 1 . . . . . . . .
## A2 . . 1 1 . . . . . .
## B2 . . 1 1 . . . . . .
## A3 . . . . 1 1 . . . .
## B3 . . . . 1 1 . . . .
## A4 . . . . . . 1 1 . .
## B4 . . . . . . 1 1 . .
## A5 . . . . . . . . 1 1
## B5 . . . . . . . . 1 1
##
## [[2]]
## 10 x 10 sparse Matrix of class "dgCMatrix"
##
## [1,] 1 . . . . . . . . .
## [2,] . 1 . . . . . . . .
## [3,] . . 1 . . . . . . .
## [4,] . . . 1 . . . . . .
## [5,] . . . . 1 . . . . .
## [6,] . . . . . 1 . . . .
## [7,] . . . . . . 1 . . .
## [8,] . . . . . . . 1 . .
## [9,] . . . . . . . . 1 .
## [10,] . . . . . . . . . 1