Insect Damages on Carrots
carrots.RdThe damage carrots data set from Phelps (1982) was used by McCullagh and Nelder (1989) in order to illustrate diagnostic techniques because of the presence of an outlier. In a soil experiment trial with three blocks, eight levels of insecticide were applied and the carrots were tested for insect damage.
data(carrots, package="robustbase")Format
A data frame with 24 observations on the following 4 variables.
- success
integer giving the number of carrots with insect damage.
- total
integer giving the total number of carrots per experimental unit.
- logdose
a numeric vector giving log(dose) values (eight different levels only).
- block
factor with levels
B1toB3
Source
Phelps, K. (1982).
Use of the complementary log-log function to describe doseresponse
relationships in insecticide evaluation field trials.
In R. Gilchrist (Ed.), Lecture Notes in Statistics, No. 14.
GLIM.82: Proceedings of the International Conference on Generalized
Linear Models; Springer-Verlag.
References
McCullagh P. and Nelder, J. A. (1989) Generalized Linear Models. London: Chapman and Hall.
Eva Cantoni and Elvezio Ronchetti (2001); JASA, and
Eva Cantoni (2004); JSS, see glmrob
Examples
data(carrots)
str(carrots)
#> 'data.frame': 24 obs. of 4 variables:
#> $ success: int 10 16 8 6 9 9 1 2 17 10 ...
#> $ total : int 35 42 50 42 35 42 32 28 38 40 ...
#> $ logdose: num 1.52 1.64 1.76 1.88 2 2.12 2.24 2.36 1.52 1.64 ...
#> $ block : Factor w/ 3 levels "B1","B2","B3": 1 1 1 1 1 1 1 1 2 2 ...
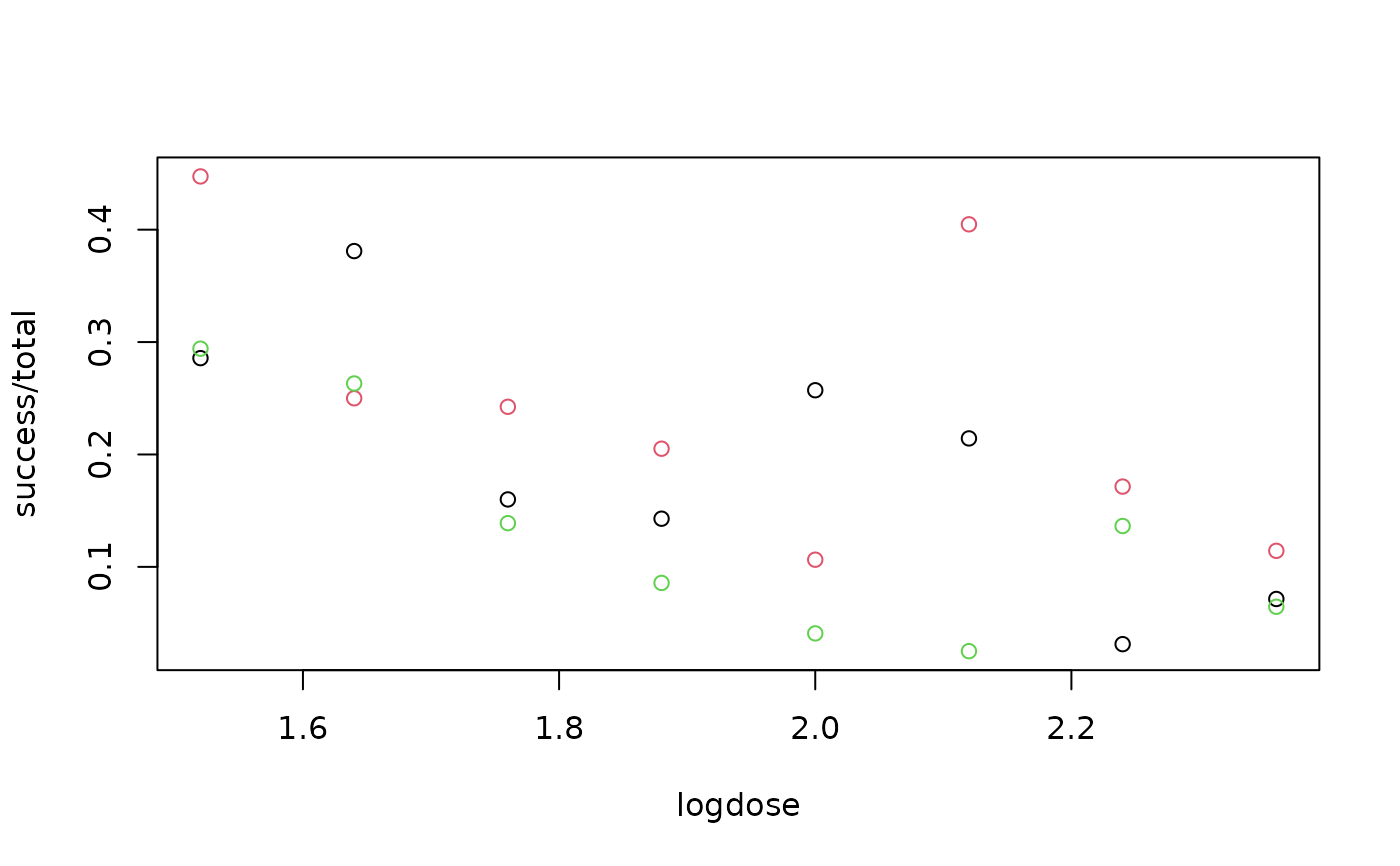
plot(success/total ~ logdose, data = carrots, col = as.integer(block))
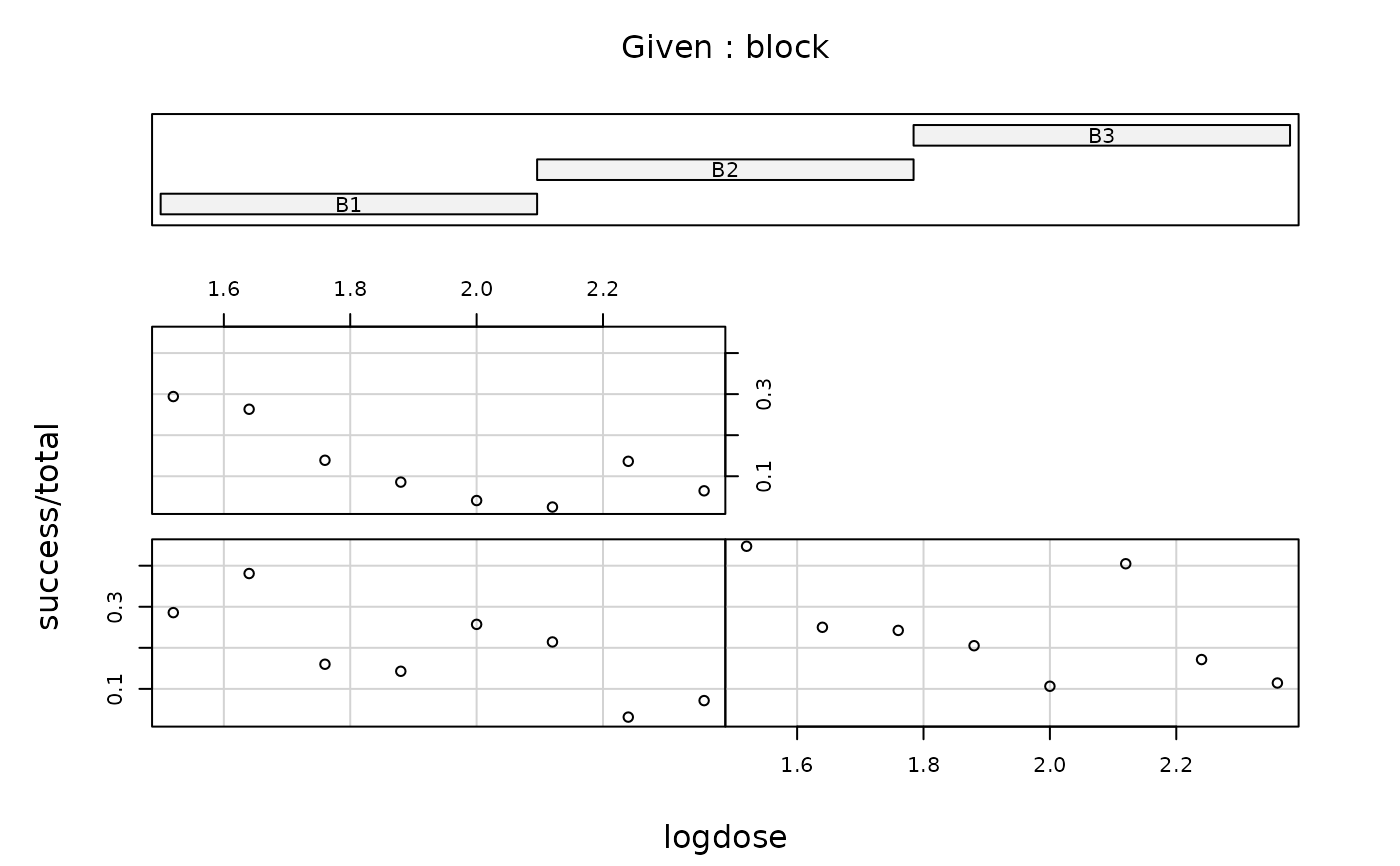
 coplot(success/total ~ logdose | block, data = carrots)
coplot(success/total ~ logdose | block, data = carrots)
 ## Classical glm
Cfit0 <- glm(cbind(success, total-success) ~ logdose + block,
data=carrots, family=binomial)
summary(Cfit0)
#>
#> Call:
#> glm(formula = cbind(success, total - success) ~ logdose + block,
#> family = binomial, data = carrots)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 2.0226 0.6501 3.111 0.00186 **
#> logdose -1.8174 0.3439 -5.285 1.26e-07 ***
#> blockB2 0.3009 0.1991 1.511 0.13073
#> blockB3 -0.5424 0.2318 -2.340 0.01929 *
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 83.344 on 23 degrees of freedom
#> Residual deviance: 39.976 on 20 degrees of freedom
#> AIC: 128.61
#>
#> Number of Fisher Scoring iterations: 4
#>
## Robust Fit (see help(glmrob)) ....
## Classical glm
Cfit0 <- glm(cbind(success, total-success) ~ logdose + block,
data=carrots, family=binomial)
summary(Cfit0)
#>
#> Call:
#> glm(formula = cbind(success, total - success) ~ logdose + block,
#> family = binomial, data = carrots)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 2.0226 0.6501 3.111 0.00186 **
#> logdose -1.8174 0.3439 -5.285 1.26e-07 ***
#> blockB2 0.3009 0.1991 1.511 0.13073
#> blockB3 -0.5424 0.2318 -2.340 0.01929 *
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 83.344 on 23 degrees of freedom
#> Residual deviance: 39.976 on 20 degrees of freedom
#> AIC: 128.61
#>
#> Number of Fisher Scoring iterations: 4
#>
## Robust Fit (see help(glmrob)) ....