Remove Columns (or Rows) From a Matrix to Make It Full Rank
fullRank.RdFrom the QR decomposition with pivoting, (qr(x, tol) if
\(n \ge p\)), if
the matrix is not of full rank, the corresponding columns (\(n \ge
p\)) or rows (\(n < p\)) are omitted to form a full rank matrix.
<!-- % -> ../R/adjoutlyingness.R -->
fullRank(x, tol = 1e-7, qrx = qr(x, tol=tol))Arguments
Value
a version of the matrix x, with less columns or rows if
x's rank was smaller than min(n,p).
If x is of full rank, it is returned unchanged.
Note
This is useful for robustness algorithms that rely on \(X\) matrices
of full rank, e.g., adjOutlyingness.
This also works for numeric data frames and whenever qr() works correctly.
See also
qr; for more sophisticated rank determination,
rankMatrix from package Matrix.
Examples
stopifnot(identical(fullRank(wood), wood))
## More sophisticated and delicate
dim(T <- tcrossprod(data.matrix(toxicity))) # 38 x 38
#> [1] 38 38
dim(T. <- fullRank(T)) # 38 x 10
#> [1] 38 10
if(requireNamespace("Matrix")) {
rMmeths <- eval(formals(Matrix::rankMatrix)$method)
rT. <- sapply(rMmeths, function(.m.) Matrix::rankMatrix(T., method = .m.))
print(rT.) # "qr" (= "qrLinpack"): 13, others rather 10
}
#> Loading required namespace: Matrix
#> tolNorm2 qr.R qrLINPACK qr useGrad maybeGrad
#> 10 10 10 10 9 10
dim(T.2 <- fullRank(T, tol = 1e-15))# 38 x 18
#> [1] 38 16
dim(T.3 <- fullRank(T, tol = 1e-12))# 38 x 13
#> [1] 38 10
dim(T.3 <- fullRank(T, tol = 1e-10))# 38 x 13
#> [1] 38 10
dim(T.3 <- fullRank(T, tol = 1e-8 ))# 38 x 12
#> [1] 38 10
dim(T.) # default from above 38 x 10
#> [1] 38 10
dim(T.3 <- fullRank(T, tol = 1e-5 ))# 38 x 10 -- still
#> [1] 38 10
plot(svd(T, 0,0)$d, log="y", main = "singular values of T", yaxt="n")
axis(2, at=10^(-14:5), las=1)
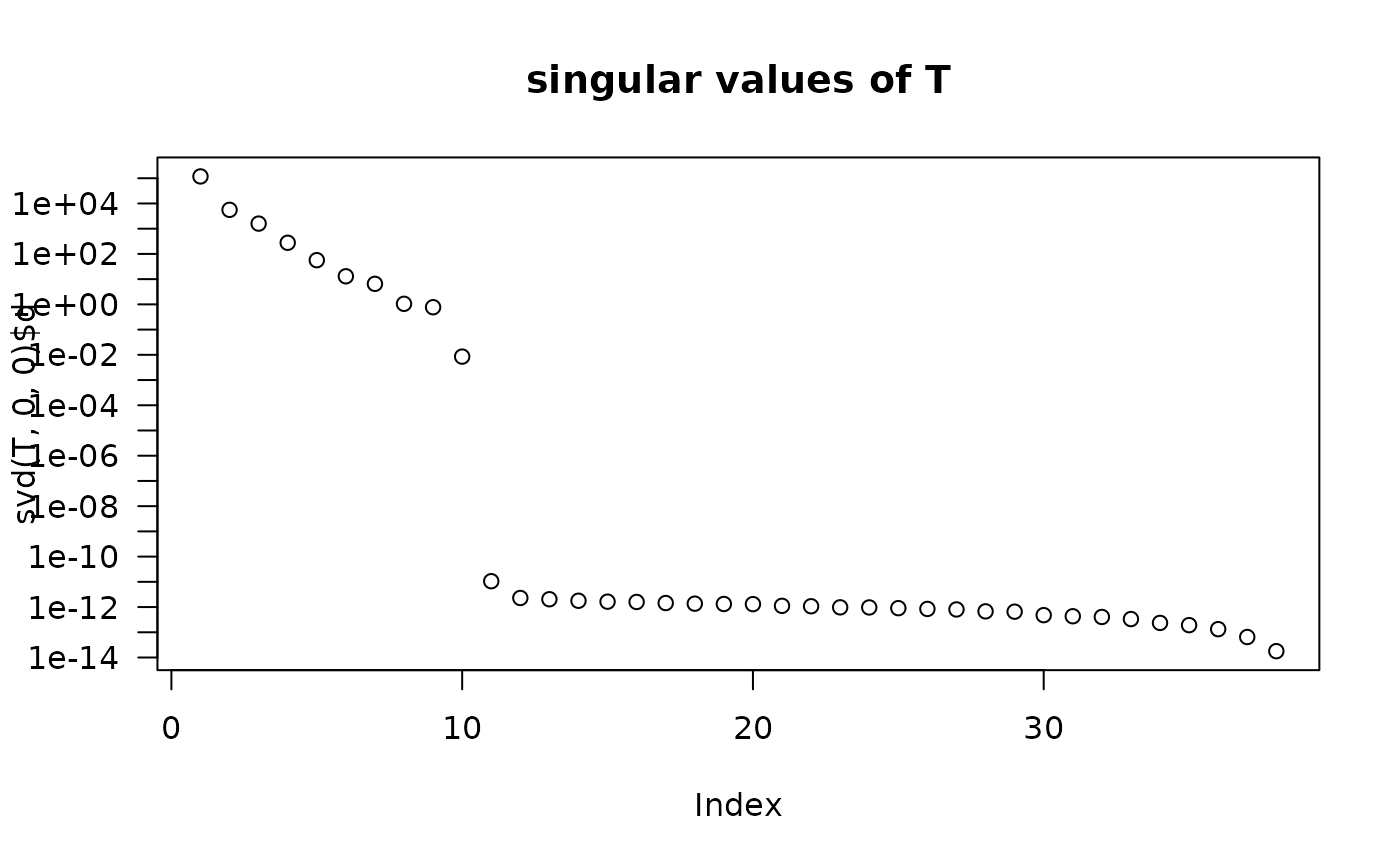 ## pretty clearly indicates that rank 10 is "correct" here.
## pretty clearly indicates that rank 10 is "correct" here.