Left and Right Medcouple, Robust Measures of Tail Weight
lmc-rmc.RdCompute the left and right ‘medcouple’, robust estimators of tail weight, in some sense robust versions of the kurtosis, the very unrobust centralized 4th moment.
Arguments
- x
a numeric vector
- mx
number, the “center” of
xwrt which the left and right parts ofxare defined:lmc(x, mx, *) := mc(x[x <= mx], *) rmc(x, mx, *) := mc(x[x >= mx], *)- na.rm
logical indicating how missing values (
NAs) should be dealt with.- doReflect
logical indicating if
mcshould also be computed on the reflected sample-x. SettingdoReflect=TRUEmakes sense for mathematical strictness reasons, as the internal MC computes the himedian() which can differ slightly from the median. Note thatmc()'s own default is true ifflength(x) <= 100.- ...
further arguments to
mc(), see its help page.
Value
each a number (unless ... contains full.result = TRUE).
References
Brys, G., Hubert, M. and Struyf, A. (2006). Robust measures of tail weight, Computational Statistics and Data Analysis 50(3), 733–759.
and those in ‘References’ of mc.
Examples
mc(1:5) # 0 for a symmetric sample
#> [1] 0
lmc(1:5) # 0
#> [1] 0
rmc(1:5) # 0
#> [1] 0
x1 <- c(1, 2, 7, 9, 10)
mc(x1) # = -1/3
#> [1] -0.3333333
c( lmc( x1), lmc( x1, doReflect=TRUE))# 0 -1/3
#> [1] 0.0000000 -0.3333333
c( rmc( x1), rmc( x1, doReflect=TRUE))# -1/3 -1/6
#> [1] -0.3333333 -0.1666667
c(-rmc(-x1), -rmc(-x1, doReflect=TRUE)) # 2/3 1/3
#> [1] 0.6666667 0.3333333
data(cushny)
lmc(cushny) # 0.2
#> [1] 0.2
rmc(cushny) # 0.45
#> [1] 0.4545455
isSym_LRmc <- function(x, tol = 1e-14)
all.equal(lmc(-x, doReflect=TRUE),
rmc( x, doReflect=TRUE), tolerance = tol)
sym <- c(-20, -5, -2:2, 5, 20)
stopifnot(exprs = {
lmc(sym) == 0.5
rmc(sym) == 0.5
isSym_LRmc(cushny)
isSym_LRmc(x1)
})
## Susceptibility to large outliers:
## "Sensitivity Curve" := empirical influence function
dX10 <- function(X) c(1:5,7,10,15,25, X) # generate skewed size-10 with 'X'
x <- c(26:40, 45, 50, 60, 75, 100)
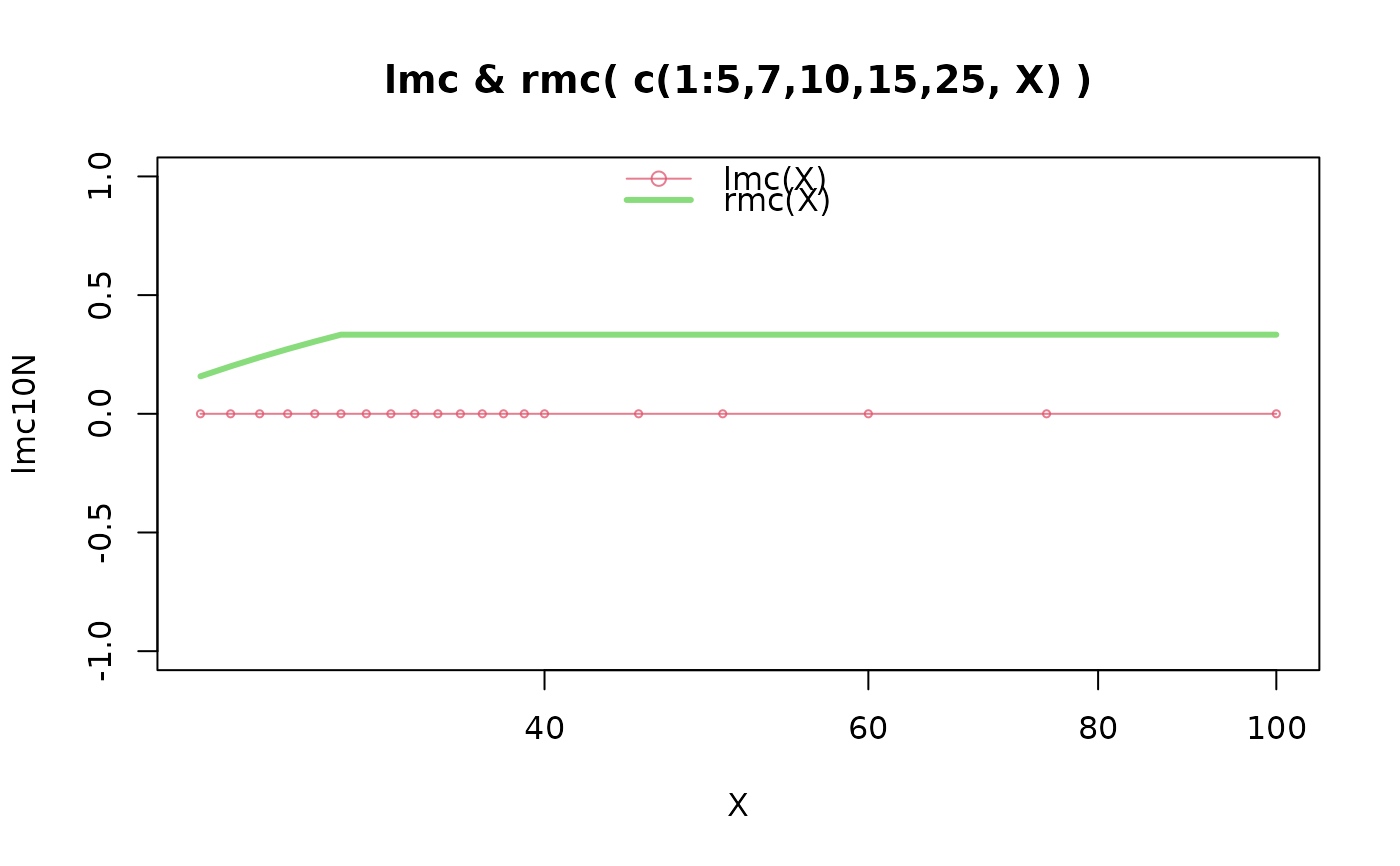
(lmc10N <- vapply(x, function(X) lmc(dX10(X)), 1))
#> [1] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
(rmc10N <- vapply(x, function(X) rmc(dX10(X)), 1))
#> [1] 0.1578947 0.2000000 0.2380952 0.2727273 0.3043478 0.3333333 0.3333333
#> [8] 0.3333333 0.3333333 0.3333333 0.3333333 0.3333333 0.3333333 0.3333333
#> [15] 0.3333333 0.3333333 0.3333333 0.3333333 0.3333333 0.3333333
cols <- adjustcolor(2:3, 3/4)
plot(x, lmc10N, type="o", cex=1/2, main = "lmc & rmc( c(1:5,7,10,15,25, X) )",
xlab=quote(X), log="x", col=cols[1])
lines(x, rmc10N, col=cols[2], lwd=3)
legend("top", paste0(c("lmc", "rmc"), "(X)"), col=cols, lty=1, lwd=c(1,3), pch = c(1, NA), bty="n")
 n <- length(x)
stopifnot(exprs = {
all.equal(current = lmc10N, target = rep(0, n))
all.equal(current = rmc10N, target = c(3/19, 1/5, 5/21, 3/11, 7/23, rep(1/3, n-5)))
## and it stays stable with outlier X --> oo :
lmc(dX10(1e300)) == 0
rmc(dX10(1e300)) == rmc10N[6]
})
n <- length(x)
stopifnot(exprs = {
all.equal(current = lmc10N, target = rep(0, n))
all.equal(current = rmc10N, target = c(3/19, 1/5, 5/21, 3/11, 7/23, rep(1/3, n-5)))
## and it stays stable with outlier X --> oo :
lmc(dX10(1e300)) == 0
rmc(dX10(1e300)) == rmc10N[6]
})