Medcouple, a Robust Measure of Skewness
mc.RdCompute the ‘medcouple’, a robust concept and estimator of skewness. The medcouple is defined as a scaled median difference of the left and right half of distribution, and hence not based on the third moment as the classical skewness.
mc(x, na.rm = FALSE, doReflect = (length(x) <= 100),
doScale = FALSE, # was hardwired=TRUE, then default=TRUE
c.huberize = 1e11, # was implicitly = Inf originally
eps1 = 1e-14, eps2 = 1e-15, # << new in 0.93-2 (2018-07..)
maxit = 100, trace.lev = 0, full.result = FALSE)Arguments
- x
a numeric vector
- na.rm
logical indicating how missing values (
NAs) should be dealt with.- doReflect
logical indicating if the internal MC should also be computed on the reflected sample
-x, with final result(mc.(x) - mc.(-x))/2. This makes sense since the internal MC,mc.()computes the himedian() which can differ slightly from the median.
- doScale
logical indicating if the internal algorithm should also scale the data (using the most distant value from the median which is unrobust and numerically dangerous); scaling has been hardwired in the original algorithm and R's
mc()till summer 2018, where it became the default. Since robustbase version 0.95-0, March 2022, the default isFALSE. As this may change the result, a message is printed about the new default, once per R session. You can suppress the message by specifyingdoScale = *explicitly, or, by settingoptions(mc_doScale_quiet=TRUE).- c.huberize
a positive number (default:
1e11) used to stabilize the sample viax <- huberize(x, c = c.huberize)for themc()computations in the case of a nearly degenerate sample (many observations practically equal to the median) or very extreme outliers. In previous versions of robustbase no such huberization was applied which is equivalent toc.huberize = Inf.- eps1, eps2
tolerance in the algorithm;
eps1is used as a for convergence tolerance, whereeps2is only used in the internalh_kern()function to prevent underflow to zero, so could be considerably smaller. The original code implicitly hard coded in Ceps1 := eps2 := 1e-13; only change with care!- maxit
maximal number of iterations; typically a few should be sufficient.
- trace.lev
integer specifying how much diagnostic output the algorithm (in C) should produce. No output by default, most output for
trace.lev = 5.- full.result
logical indicating if the full return values (from C) should be returned as a list via
attr(*, "mcComp").
Value
a number between -1 and 1, which is the medcouple, \(MC(x)\).
For r <- mc(x, full.result = TRUE, ....), then
attr(r, "mcComp") is a list with components
- medc
the medcouple \(mc.(x)\).
- medc2
the medcouple \(mc.(-x)\) if
doReflect=TRUE.- eps
tolerances used.
- iter,iter2
number of iterations used.
- converged,converged2
logical specifying “convergence”.
Convergence Problems
For extreme cases there were convergence problems which should not
happen anymore as we now use doScale=FALSE and huberization (when
c.huberize < Inf).
The original algorithm and mc(*, doScale=TRUE) not only centers
the data around the median but
also scales them by the extremes which may have a negative effect
e.g., when changing an extreme outlier to even more extreme, the
result changes wrongly; see the 'mc10x' example.
References
Guy Brys, Mia Hubert and Anja Struyf (2004) A Robust Measure of Skewness; JCGS 13 (4), 996–1017.
Hubert, M. and Vandervieren, E. (2008). An adjusted boxplot for skewed distributions, Computational Statistics and Data Analysis 52, 5186–5201.
Lukas Graz (2021). Improvement of the Algorithms for the Medcoule and the Adjusted Outlyingness; unpublished BSc thesis, supervised by M.Maechler, ETH Zurich.
See also
Qn for a robust measure of scale (aka
“dispersion”), ....
Examples
mc(1:5) # 0 for a symmetric sample
#> [1] 0
x1 <- c(1, 2, 7, 9, 10)
mc(x1) # = -1/3
#> [1] -0.3333333
data(cushny)
mc(cushny) # 0.125
#> [1] 0.125
stopifnot(mc(c(-20, -5, -2:2, 5, 20)) == 0,
mc(x1, doReflect=FALSE) == -mc(-x1, doReflect=FALSE),
all.equal(mc(x1, doReflect=FALSE), -1/3, tolerance = 1e-12))
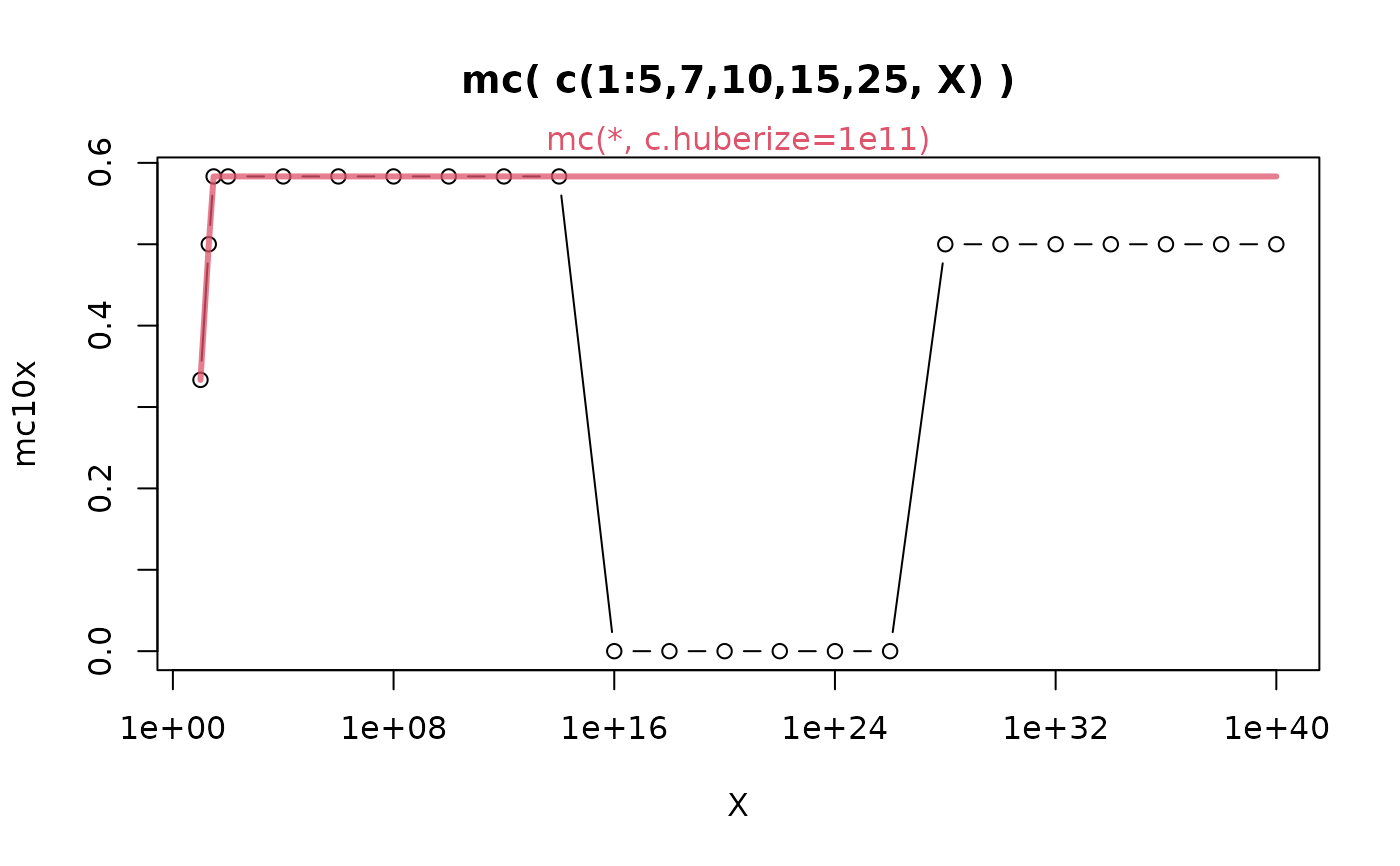
## Susceptibility of the current algorithm to large outliers :
dX10 <- function(X) c(1:5,7,10,15,25, X) # generate skewed size-10 with 'X'
x <- c(10,20,30, 100^(1:20))
## (doScale=TRUE, c.huberize=Inf) were (implicit) defaults in earlier {robustbase}:
(mc10x <- vapply(x, function(X) mc(dX10(X), doScale=TRUE, c.huberize=Inf), 1))
#> [1] 0.3333333 0.5000000 0.5833333 0.5833333 0.5833333 0.5833333 0.5833333
#> [8] 0.5833333 0.5833333 0.5833333 0.0000000 0.0000000 0.0000000 0.0000000
#> [15] 0.0000000 0.0000000 0.5000000 0.5000000 0.5000000 0.5000000 0.5000000
#> [22] 0.5000000 0.5000000
## limit X -> Inf should be 7/12 = 0.58333... but that "breaks down a bit" :
plot(x, mc10x, type="b", main = "mc( c(1:5,7,10,15,25, X) )", xlab="X", log="x")
## The new behavior is much preferable {shows message about new 'doScale=FALSE'}:
(mc10N <- vapply(x, function(X) mc(dX10(X)), 1))
#> [1] 0.3333333 0.5000000 0.5833333 0.5833333 0.5833333 0.5833333 0.5833333
#> [8] 0.5833333 0.5833333 0.5833333 0.5833333 0.5833333 0.5833333 0.5833333
#> [15] 0.5833333 0.5833333 0.5833333 0.5833333 0.5833333 0.5833333 0.5833333
#> [22] 0.5833333 0.5833333
lines(x, mc10N, col=adjustcolor(2, 3/4), lwd=3)
mtext("mc(*, c.huberize=1e11)", col=2)
 stopifnot(all.equal(c(4, 6, rep(7, length(x)-2))/12, mc10N))
## Here, huberization already solves the issue:
mc10NS <- vapply(x, function(X) mc(dX10(X), doScale=TRUE), 1)
stopifnot(all.equal(mc10N, mc10NS))
stopifnot(all.equal(c(4, 6, rep(7, length(x)-2))/12, mc10N))
## Here, huberization already solves the issue:
mc10NS <- vapply(x, function(X) mc(dX10(X), doScale=TRUE), 1)
stopifnot(all.equal(mc10N, mc10NS))