Robust Regression Outlier Statistics
outlierStats.RdSimple statistics about observations with robustness weight of almost zero for models that include factor terms. The number of rejected observations and the mean robustness weights are computed for each level of each factor included in the model.
outlierStats(object, x = object$x, control = object$control
, epsw = control$eps.outlier
, epsx = control$eps.x
, warn.limit.reject = control$warn.limit.reject
, warn.limit.meanrw = control$warn.limit.meanrw
, shout = NA)Arguments
- object
object of class
"lmrob", typically the result of a call tolmrob.- x
design matrix
- control
list as returned by
lmrob.control().- epsw
limit on the robustness weight below which an observation is considered to be an outlier. Either a
numeric(1)or afunctionthat takes the number of observations as an argument.- epsx
limit on the absolute value of the elements of the design matrix below which an element is considered zero. Either a numeric(1) or a function that takes the maximum absolute value in the design matrix as an argument.
- warn.limit.reject
limit of ratio \(\#\mbox{rejected} / \#\mbox{obs in level}\) above (\(\geq\)) which a warning is produced. Set to
NULLto disable warning.- warn.limit.meanrw
limit of the mean robustness per factor level below which (\(\leq\)) a warning is produced. Set to
NULLto disable warning.- shout
a
logical(scalar) indicating if large"Ratio"or small"Mean.RobWeight"should lead to correspondingwarning()s; cutoffs are determined bywarn.limit.rejectandwarn.limit.meanrw, above. By default,NA; setting it toFALSEorTRUEdisables or unconditionally enables “shouting”.
Details
For models that include factors, the fast S-algorithm used by
lmrob can produce “bad” fits for some of the
factor levels, especially if there are many levels with only a
few observations. Such a “bad” fit is characterized as a
fit where most of the observations in a level of a factor are
rejected, i.e., are assigned robustness weights of zero or nearly
zero. We call such a fit a “local exact fit”.
If a local exact fit is detected, then we recommend to increase some
of the control parameters of the “fast S”-algorithm. As a first
aid solution in such cases, one can use setting="KS2014", see also
lmrob.control.
This function is called internally by lmrob to issue a
warning if a local exact fit is detected. The output is available as
ostats in objects of class "lmrob" (only if the statistic
is computed).
Value
A data frame for each column with any zero elements as well as an
overall statistic. The data frame consist of the names of the
coefficients in question, the number of non-zero observations in that
level (N.nonzero), the number of rejected observations
(N.rejected), the ratio of rejected observations to the
number of observations in that level (Ratio) and the mean
robustness weight of all the observations in the corresponding level
(Mean.RobWeight).
References
Koller, M. and Stahel, W.A. (2017) Nonsingular subsampling for regression S estimators with categorical predictors, Computational Statistics 32(2): 631–646. doi:10.1007/s00180-016-0679-x
See also
lmrob.control for the default values of the control
parameters; summarizeRobWeights.
Examples
## artificial data example
data <- expand.grid(grp1 = letters[1:5], grp2 = letters[1:5], rep=1:3)
set.seed(101)
data$y <- c(rt(nrow(data), 1))
## compute outlier statistics for all the estimators
control <- lmrob.control(method = "SMDM",
compute.outlier.stats = c("S", "MM", "SMD", "SMDM"))
## warning is only issued for some seeds
set.seed(2)
fit1 <- lmrob(y ~ grp1*grp2, data, control = control)
#> Warning: Detected possible local breakdown of S-estimate in coefficient 'grp1c:grp2c'.
#> Use lmrob argument 'setting="KS2014"' to avoid this problem.
## do as suggested:
fit2 <- lmrob(y ~ grp1*grp2, data, setting = "KS2014")
## the plot function should work for such models as well
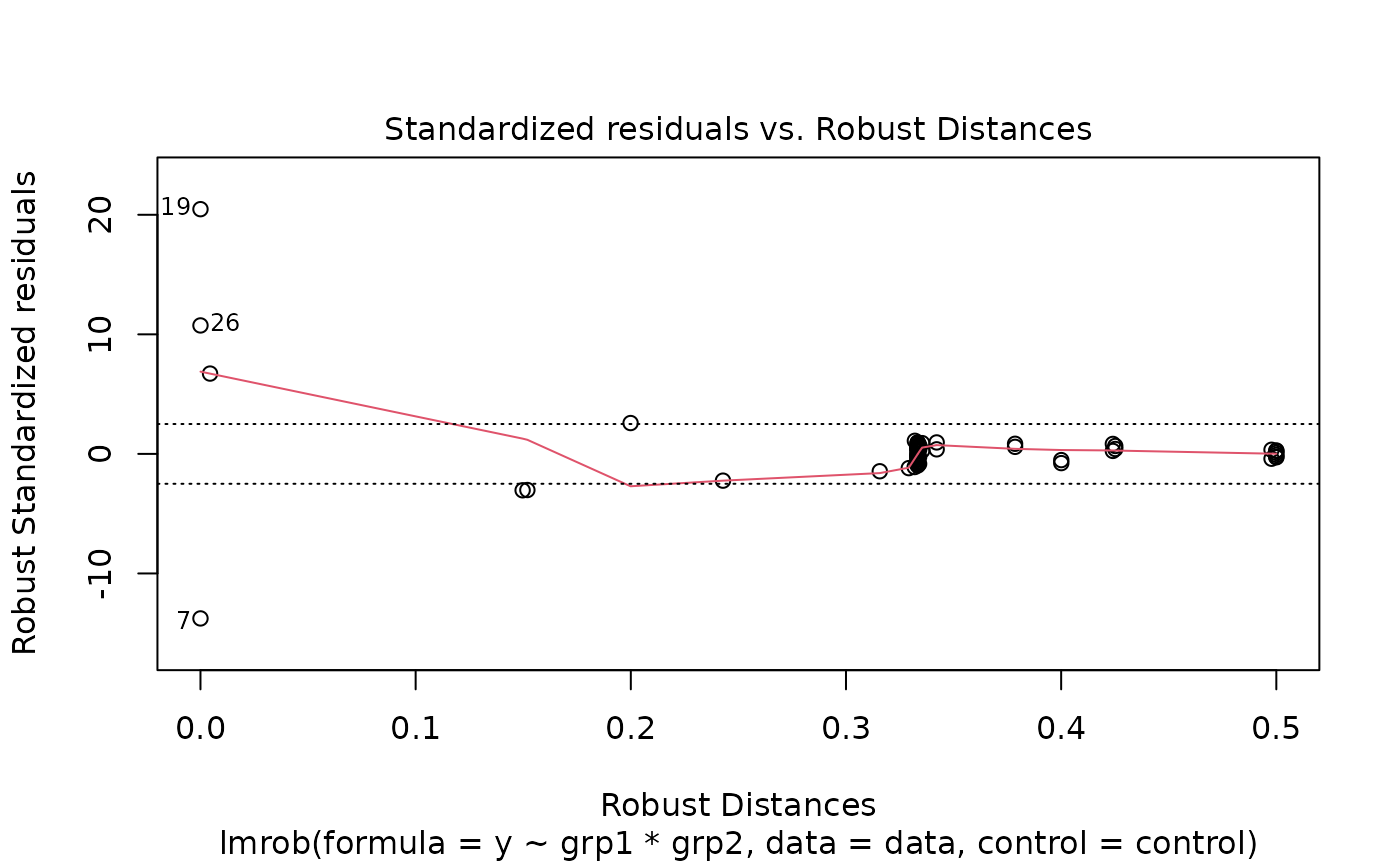
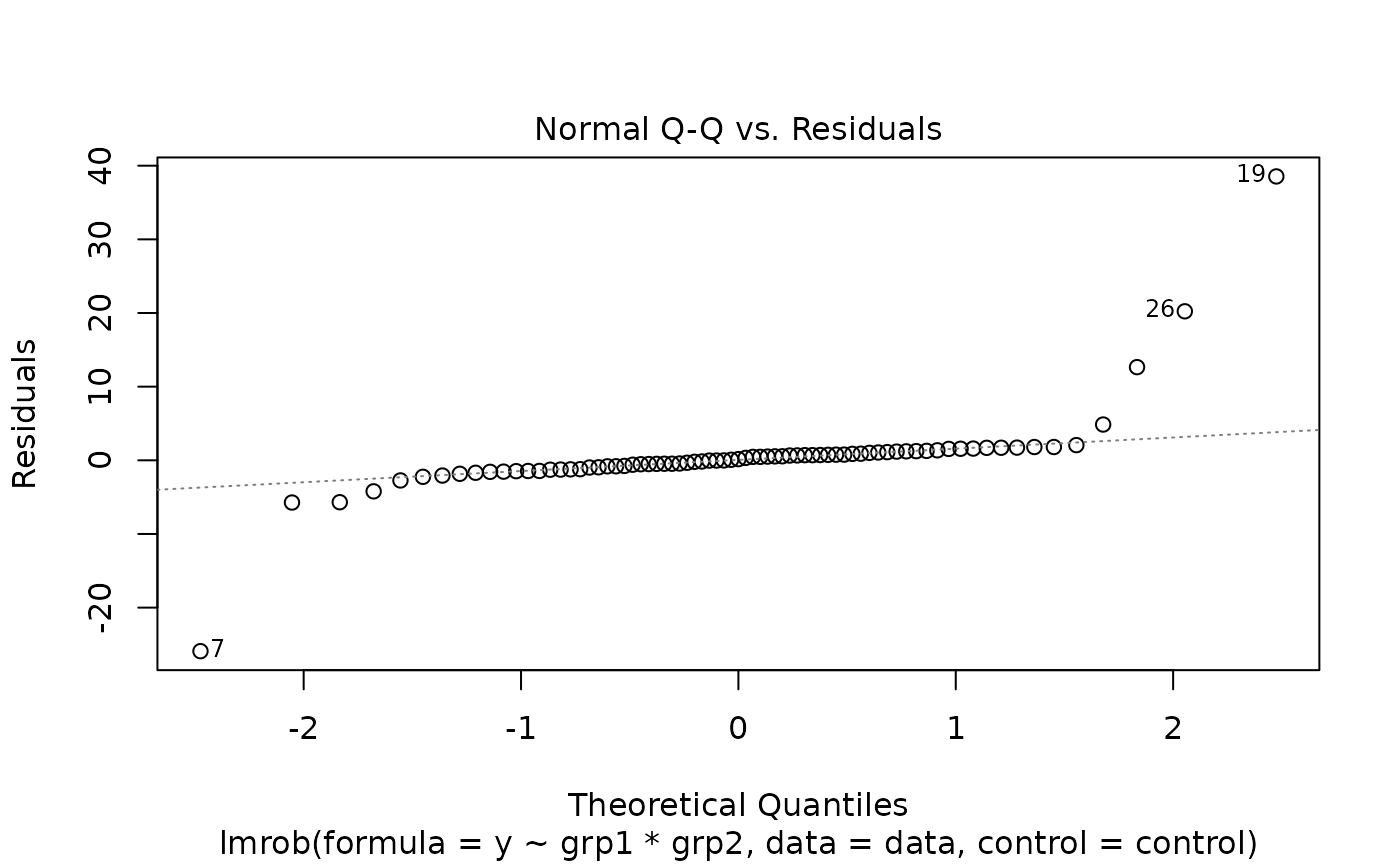
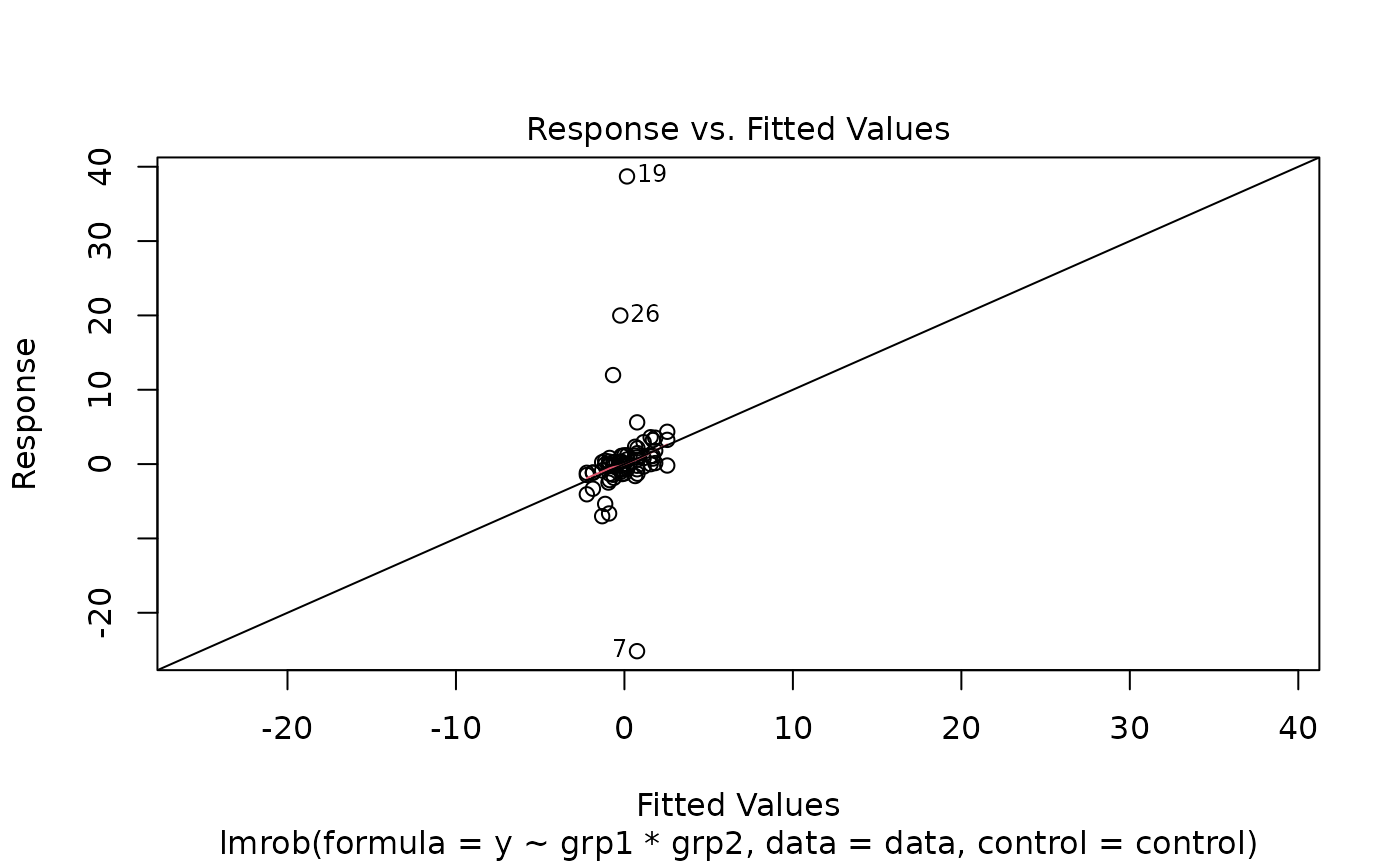
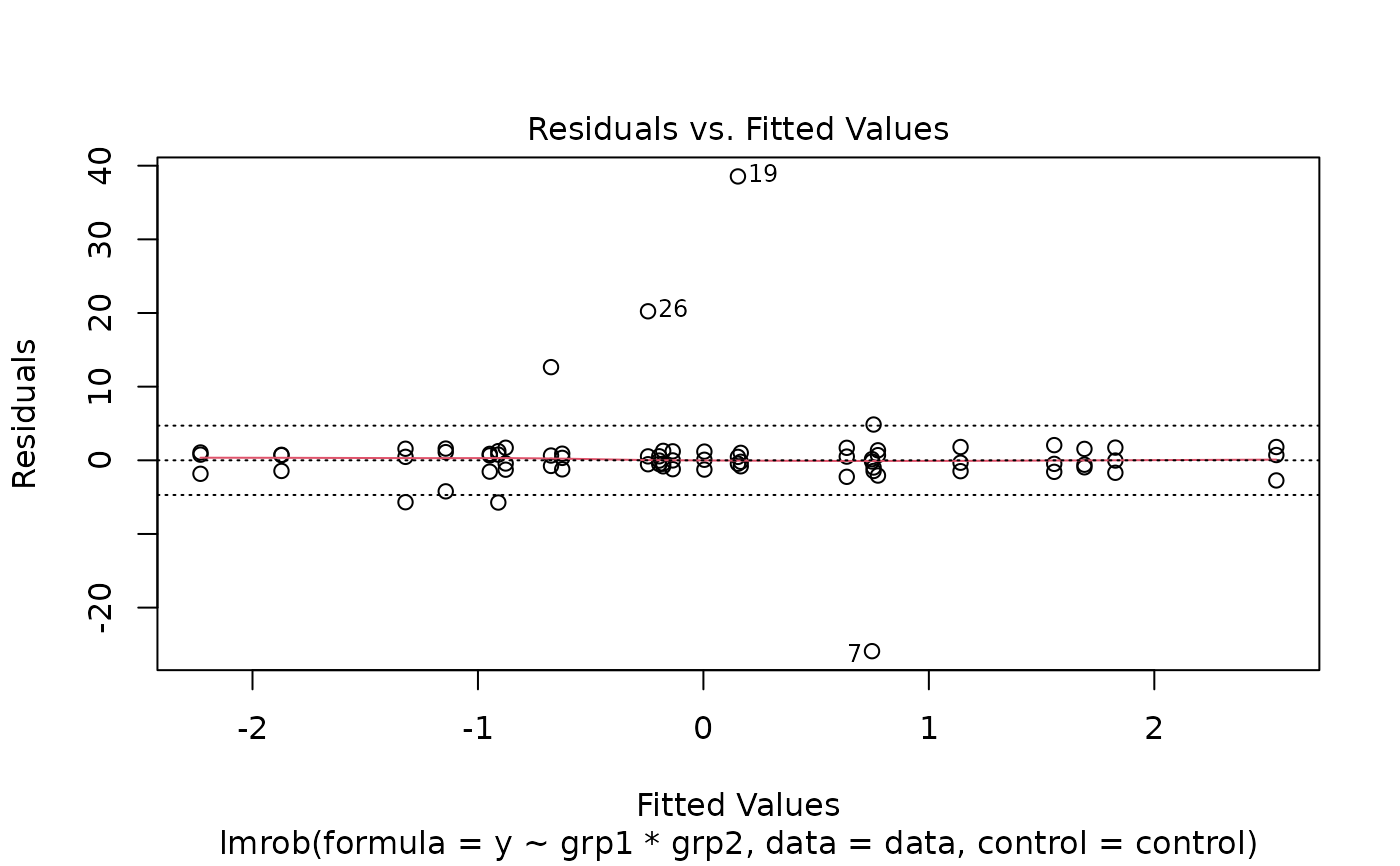
plot(fit1)
#> recomputing robust Mahalanobis distances
#> Warning: Failed to compute robust Mahalanobis distances, reverting to robust leverages.
 if (FALSE) { # \dontrun{
## access statistics:
fit1$ostats ## SMDM
fit1$init$ostats ## SMD
fit1$init$init$ostats ## SM
fit1$init$init$init.S$ostats ## S
} # }
if (FALSE) { # \dontrun{
## access statistics:
fit1$ostats ## SMDM
fit1$init$ostats ## SMD
fit1$init$init$ostats ## SM
fit1$init$init$init.S$ostats ## S
} # }