Integrating User Defined Functions into rxode2
Source:vignettes/articles/Integrating-User-Defined-Functions-into-rxode2.Rmd
Integrating-User-Defined-Functions-into-rxode2.RmdUser Defined Functions
library(rxode2)
#> rxode2 3.0.3 using 2 threads (see ?getRxThreads)
#> no cache: create with `rxCreateCache()`When defining models you may have wished to write a small R function
or make a function integrate into rxode2 somehow. This
article discusses 4 ways to do this:
A R-based user function which can be loaded as a simple function or in certain circumstances translated to C to run more efficiently
A C function that you define and integrate into code
A user defined function that changes
rxode2ui code by replacing the function withrxode2code. This can be in the presence or absence of the data for simulation or estimation.
R based user functions
A R-based user function is the most convenient to include in the ODE,
but is slower than what you could have done if it was written in
C , C++ or some other compiled language. This
was requested in
github with an appropriate example; However, I will use a very
simple example here to simply illustrate the concepts.
newAbs <- function(x) {
if (x < 0) {
-x
} else {
x
}
}
f <- rxode2({
a <- newAbs(time)
})
#> using C compiler: 'gcc (Ubuntu 13.3.0-6ubuntu2~24.04) 13.3.0'
e <- et(-10, 10, length.out=40)Now that the ODE has been compiled the R functions will be called while solving the ODE. Since this is calling R, this forces the parallization to be turned off since R is single-threaded. It also takes more time to solve since it is shuttling back and forth between R and C. Lets see how this very simple function performs:
mb1 <- microbenchmark::microbenchmark(withoutC=suppressWarnings(rxSolve(f,e)))
library(ggplot2)
autoplot(mb1) + rxTheme()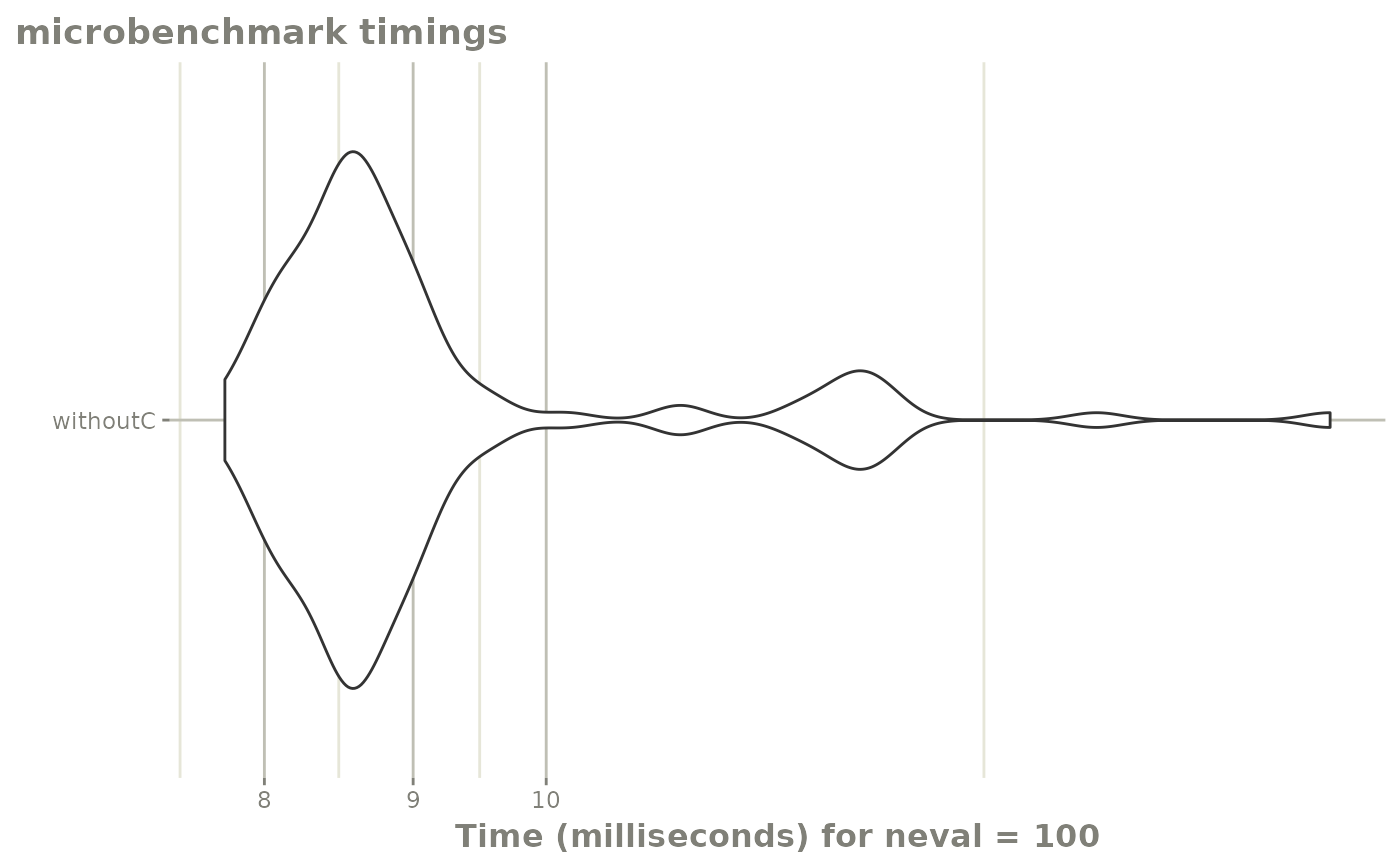
Not terribly bad, even though it is shuffling between R and C.
You can make it a better by converting the functions to C:
# Create C functions automatically with `rxFun()`
rxFun(newAbs)
#> > finding duplicate expressions in d(newAbs)/d(x)...
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> > optimizing duplicate expressions in d(newAbs)/d(x)...
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> converted R function 'newAbs' to C (will now use in rxode2)
#> converted R function 'rx_newAbs_d_x' to C (will now use in rxode2)
#> Added derivative table for 'newAbs'
# Recompile to use the C functions
# Note it would recompile anyway if you didn't do this step,
# it just makes sure that it doesn't recompile every step in
# the benchmark
f <- rxode2({
a <- newAbs(time)
})
#> using C compiler: 'gcc (Ubuntu 13.3.0-6ubuntu2~24.04) 13.3.0'
mb2 <- microbenchmark::microbenchmark(withC=rxSolve(f,e, cores=1))
mb <- rbind(mb1, mb2)
autoplot(mb) + rxTheme() + xgxr::xgx_scale_y_log10()
#> Scale for y is already present.
#> Adding another scale for y, which will replace the existing scale.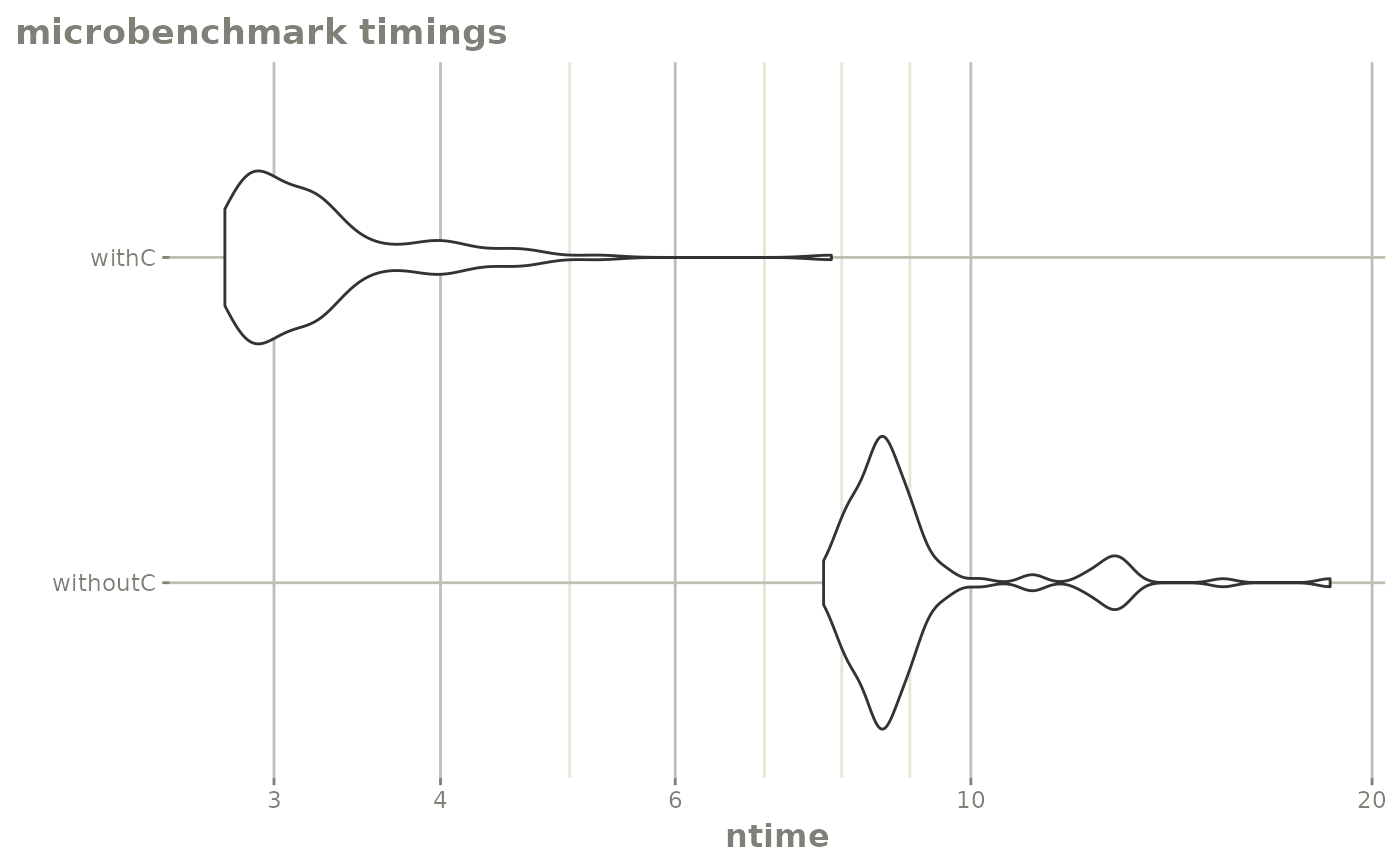
print(mb)
#> Unit: milliseconds
#> expr min lq mean median uq max neval
#> withoutC 7.753618 8.362809 9.228476 8.626327 9.058295 18.602100 100
#> withC 2.755867 2.903652 3.299911 3.099551 3.358154 7.858736 100The C version is almost twice as fast as the R version. You may have
noticed the conversion also created C versions of the first derivative.
This is done automatically and gives not just C versions of function,
but C versions of the derivatives and registers them with
rxode2. This allows the C versions to work with not only
rxode2 but nlmixr2 models.
This function was setup in advance to allow this type of conversion.
In general the derivatives will be calculated if there is not a
return() statement in the user defined function. This means
simply let R return the last value instead of explictly calling out the
return(). Many people prefer this method of coding.
Even if there is a return function, the function could
be converted to C. In the github issue, they used a
function that would not convert the derivatives:
# Light
f_R <- function(actRad, k_0, a_k) {
photfac <- a_k * actRad + k_0
if (photfac > 1) {
photfac = 1
}
return(photfac)
}
rxFun(f_R)
#> function contains return statement; derivatives not calculated
#> converted R function 'f_R' to C (will now use in rxode2)While this is still helpful because some functions have early
returns, the nlmixr2 models requiring derivatives would be
calculated be non-optimized finite differences when this occurs. While
this gets into the internals of rxode2 and
nlmixr2 you can see this more easily when calculating the
derivatives:
rxFromSE("Derivative(f_R(actRad, k_0, a_k),k_0)")
#> [1] "(f_R(actRad,(k_0)+6.05545445239334e-06,a_k)-f_R(actRad,k_0,a_k))/6.05545445239334e-06"Whereas the originally defined function newAbs() would
use the new derivatives calculated as well:
rxFromSE("Derivative(newAbs(x),x)")
#> [1] "rx_newAbs_d_x(x)"In some circumstances, the conversion to C is not possible, though you can still use the R function.
There are some requirements for R functions to be integrated into the rxode2 system:
The function must have a set number of arguments, variable arguments like
f(…)are currently not allowed.The function is given each argument as a single number, and the function should return a single number
If these requirements are met you can use the R function in rxode2. Additional requirements for conversion to C include:
-
Any functions that you use within the R function must be understood and available to
rxode2.- Practically speaking if you have
fun2()which refers tofun1(),fun1()must be changed to C code and available torxode2before changing the functionfun2()to C.
- Practically speaking if you have
The functions can include
if/elseassignments or simple return statements (either by returning a value or having that value on a line by itself). Special R control structures and functions (likeforandlapply) cannot be present.The function cannot refer to any package functions
As mentioned, if the
return()statement is present, the derivative C functions andrxode2’s derivative table is not updated.
C based functions
You can add your own C functions directly into rxode2 as well using
rxFun():
fun <- "
double fun(double a, double b, double c) {
return a*a+b*a+c;
}
" ## C-code for function
rxFun("fun", c("a", "b", "c"), fun)If you wanted you could also use C functions or expressions for the
derivatives by using the rxD() function:
rxD("fun", list(
function(a, b, c) { # derivative of arg1: a
paste0("2*", a, "+", b)
},
function(a, b, c) { # derivative of arg2: b
return(a)
},
function(a, b, c) { # derivative of arg3: c
return("0.0")
}
))Removing the function with rxRmFun() will also remove
the derivative table:
rxRmFun("fun")Functions to insert rxode2 code into the current
model
This replaces rxode2 code in the current model with some
expansion. This can allow more R-like functions inside of the rxode2 ui
models, as well as adding approximating functions like polynomials,
splines and neural networks.
An example that allows more R-like functions is
below:
f <- function() {
model({
a <- rxpois(lambda=lam)
})
}
# Which will evaluate into a standard rxode2 function that does not
# support named arguments (since it is translated to C)
f()
# Which is still true in the standard rxode2:
try(rxode2({
a <- rxpois(lambda=lam)
}))This is accomplished by a combination of two functions, which are highly commented:
rxUdfUi.rxpois <- function(fun) {
# Fun is the language object (ie quoted R object) to be evaluated or
# changed in the code
.fun <- fun
# Since the `rxpois` function is built into the rxode2 we need to
# have a function with a different conflicts. In this case, I take
# the function name (fun[[1]]), and prepend a ".", which follows
# `rxode2`'s naming convention of un-exported functions.
#
# This next evaluation changes the expression function to .rxpois()
.fun[[1]] <- str2lang(paste0(".", deparse1(fun[[1]])))
# Since this is still a R expression, you can then evaluate the
# function .rxpois to produce the proper code:
eval(.fun)
}
# The above s3 method can be registered in a package or you can use
# the following code to register it in your session:
rxode2::.s3register("rxode2::rxUdfUi", "rxpois")
# This is the function that changes the code as needed
.rxpois <- function(lambda) {
# The first part of this code tries to change the value into a
# character. This handles cases like rxpois(lambda=lam),
# rxpois(lam), rxpois("lam"). It also tries to evaluate the
# argument supplied to lambda in case it comes from a different
# location.
.lam <- as.character(substitute(lambda))
.tmp <- try(force(lambda), silent=TRUE)
if (!inherits(.tmp, "try-error")) {
if (is.character(.tmp)) {
.lam <- lambda
}
}
# This part creates a list with the replacement text, in this case
# it woulb be rxpois(lam) where there is no equals included, as
# required by `rxode2`:
list(replace=paste0("rxpois(", .lam, ")"))
}In general the list that the function needs to return can have:
$replace– The text that will be replaced$before– lines that will be placed in the model before the current function is found$after– lines that are added in the model after the current function is found$iniDf– the initial estimatesdata.framefor this problem. While calling this function, you can retrieve the initial conditions currently used parsing you can get the prior value withrxUdfUiIniDf()and then you can modify it inside the function and return the newdata.framein this list element. This allows you add/delete initial estimates from the model as well as modify the model lines themselves.$uiUseData– whenTRUE, this instructsrxode2andnlmixr2estto re-parse this function in the presence of data, this means a bit more function setup will need to be done.$uiUseMv– whenTRUEthis instructsrxode2to re-parse the function after the initial model variables are calculated.
In addition to the rxUdfUiIniDf() you can get
information about the parser:
rxUdfUiParsing()returns if the rxode2 ui function is being parsed currently (this allows a function to be overloaded as audffor calling fromrxode2as well as a function for modifying the model).rxUdfUiNum()during parsing the function you are calling (in the example aboverxpois()can be called multiple times. This gives the number of the function in the model in order (the first would give1the second,2, etc). This can be used to create unique variables with functions likerxIntToLetter()orrxIntToBase().rxUdfUiIniLhs()which gives the left-handed side of the equation where the function is found. This is also aRlanguage object.rxUdfUiIniMv()gives the model variables for parsing (can be used in functions likelinCmt())rxUdfUiData()which specifies the data that are being used to simulate, estimate, etc.rxUdfUiEst()which gives the estimation/simulation method that is being used with the model. For example, with simulation it would berxSolve.
Using model variables in rxode2 ui models
You can also take and change the model and take into consideration
the rxode2 model variables before the full ui
has completed its parsing. These rxode2 model variables has
information that might change what variables you make or names of
variables. For example it has what is on the left hand side of the
equations ($lhs), what are the input parameters
($params) and what is the ODE states
($state)).
If you are using this approach, you will likely need to do the following steps:
When data are not being processed, you need to put the function in an
rxode2acceptable form, no named arguments, no strings, and only numbers or variables in the output.The number of arguments of this output needs to be declared in the
S3method by adding the attribute"nargs"to method. For example, the built intestMod1()ui modification function uses only one argument when parsed
Below is a commented example of the model variables example:
testMod1 <- function(val=1) {
# This converts the val to a character if it is somthing like testMod1(b)
.val <- as.character(substitute(val))
.tmp <- suppressWarnings(try(force(val), silent = TRUE))
if (!inherits(.tmp, "try-error")) {
if (is.character(.tmp)) {
.val <- val
}
}
# This does the UI parsing
if (rxUdfUiParsing()) {
# See if the model variables are available
.mv <- rxUdfUiMv()
if (is.null(.mv)) {
# Put this in a rxode2 low level acceptible form, no complex
# expressions, no named arguments, something that is suitable
# for C.
#
# The `uiUsMv` tells the parser this needs to be reparsed when
# the model variables become avaialble during parsing.
return(list(replace=paste0("testMod1(", .val, ")"),
uiUseMv=TRUE))
} else {
# Now that we have the model variables, we can then do something
# about this
.vars <- .mv$params
if (length(.vars) > 0) {
# If there is parameters available, this dummy function times
# the first input function by the value specified
return(list(replace=paste0(.vars[1], "*", .val)))
} else {
# If the value isn't availble, simply replace the function
# with the value.
return(list(replace=.val))
}
}
}
stop("This function is only for use in rxode2 ui models",
call.=FALSE)
}
rxUdfUi.testMod1 <- function(fun) {
eval(fun)
}
# To allow this to go to the next step, you need to declare how many
# arguments this argument has, in this case 1. Bu adding the
# attribute "nargs", rxode2 lower level parser knows how to handle
# this new function. This allows rxode2 to generate the model
# variables and send it to the next step.
attr(rxUdfUi.testMod1, "nargs") <- 1L
# If you are in a package, you can use the rxoygen tag @export to
# register this as a rxode2 model definition.
#
# If you are using this in your own script, you need to register the s3 function
# One way to do this is:
rxode2::.s3register("rxode2::rxUdfUi", "testMod1")
## These are some examples of this function in use:
f <- function() {
model({
a <- b + testMod1(3)
})
}
f <- f()
print(f)
#> -- rxode2-based Pred model -----------------------------------------------------
#> -- Model (Normalized Syntax): --
#> function() {
#> model({
#> a <- b + (b * 3)
#> })
#> }
f <- function() {
model({
a <- testMod1(c)
})
}
f <- f()
print(f)
#> -- rxode2-based Pred model -----------------------------------------------------
#> -- Model (Normalized Syntax): --
#> function() {
#> model({
#> a <- (c * c)
#> })
#> }
f <- function() {
model({
a <- testMod1(1)
})
}
f <- f()
print(f)
#> -- rxode2-based Pred model -----------------------------------------------------
#> -- Model (Normalized Syntax): --
#> function() {
#> model({
#> a <- 1
#> })
#> }Using data for rxode2 ui modification models
The same steps are needed to use the data in the model replacement;
You can then use the data and the model to replace the values inside the
model. A worked example linMod() is included that has the
ability to use:
- model variables,
- put lines before or after the model,
- add initial conditions
- And use data in the initial estimates
# You can print the code:
linMod
#> function (variable, power, dv = "dv", intercept = TRUE, type = c("replace",
#> "before", "after"), num = NULL, iniDf = NULL, data = FALSE,
#> mv = FALSE)
#> {
#> .dv <- as.character(substitute(dv))
#> .tmp <- suppressWarnings(try(force(dv), silent = TRUE))
#> if (!inherits(.tmp, "try-error")) {
#> if (is.character(.tmp)) {
#> .dv <- dv
#> }
#> }
#> .var <- as.character(substitute(variable))
#> .tmp <- try(force(variable), silent = TRUE)
#> .doExp3 <- FALSE
#> if (!inherits(.tmp, "try-error")) {
#> if (is.character(.tmp)) {
#> .var <- variable
#> }
#> else if (!inherits(.tmp, "formula")) {
#> .dv <- as.character(substitute(dv))
#> .tmp <- suppressWarnings(try(force(dv), silent = TRUE))
#> if (!inherits(.tmp, "try-error")) {
#> if (is.character(.tmp)) {
#> .dv <- dv
#> }
#> }
#> }
#> else if (length(variable) == 2) {
#> if (!identical(variable[[1]], quote(`~`))) {
#> stop("unexpected formula, needs to be the form ~x^3",
#> call. = FALSE)
#> }
#> .doExp3 <- TRUE
#> .exp3 <- variable[[2]]
#> }
#> else {
#> if (length(variable) != 3) {
#> stop("unexpected formula, needs to be the form dv~x^3",
#> call. = FALSE)
#> }
#> if (!identical(variable[[1]], quote(`~`))) {
#> stop("unexpected formula, needs to be the form dv~x^3",
#> call. = FALSE)
#> }
#> .dv <- as.character(variable[[2]])
#> data <- TRUE
#> .exp3 <- variable[[3]]
#> .doExp3 <- TRUE
#> }
#> if (.doExp3) {
#> if (length(.exp3) == 1) {
#> .var <- variable <- as.character(.exp3)
#> power <- 1
#> }
#> else if (length(.exp3) == 3) {
#> if (!identical(.exp3[[1]], quote(`^`))) {
#> stop("unexpected formula, needs to be the form dv~x^3",
#> call. = FALSE)
#> }
#> if (!is.numeric(.exp3[[3]])) {
#> stop("unexpected formula, needs to be the form dv~x^3",
#> call. = FALSE)
#> }
#> .var <- variable <- as.character(.exp3[[2]])
#> power <- .exp3[[3]]
#> }
#> else {
#> stop("unexpected formula, needs to be the form dv~x^3",
#> call. = FALSE)
#> }
#> }
#> }
#> checkmate::assertCharacter(.var, len = 1L, any.missing = FALSE,
#> pattern = "^[.]*[a-zA-Z]+[a-zA-Z0-9._]*$", min.chars = 1L,
#> .var.name = "variable")
#> checkmate::assertCharacter(.dv, len = 1L, any.missing = FALSE,
#> pattern = "^[.]*[a-zA-Z]+[a-zA-Z0-9._]*$", min.chars = 1L,
#> .var.name = "dv")
#> checkmate::assertLogical(intercept, len = 1L, any.missing = FALSE)
#> checkmate::assertIntegerish(power, lower = ifelse(intercept,
#> 0L, 1L), len = 1L)
#> if (is.null(num)) {
#> num <- rxUdfUiNum()
#> }
#> checkmate::assertIntegerish(num, lower = 1, any.missing = FALSE,
#> len = 1)
#> if (mv && is.null(rxUdfUiMv())) {
#> if (intercept) {
#> return(list(replace = paste0("linModM(", .var, ", ",
#> power, ")"), uiUseMv = TRUE))
#> }
#> else {
#> return(list(replace = paste0("linModM0(", .var, ", ",
#> power, ")"), uiUseMv = TRUE))
#> }
#> }
#> if (data && is.null(rxUdfUiData())) {
#> if (intercept) {
#> return(list(replace = paste0("linModD(", .var, ", ",
#> power, ", ", .dv, ")"), uiUseData = TRUE))
#> }
#> else {
#> return(list(replace = paste0("linModD0(", .var, ", ",
#> power, ",", .dv, ")"), uiUseData = TRUE))
#> }
#> }
#> if (is.null(iniDf)) {
#> iniDf <- rxUdfUiIniDf()
#> }
#> assertIniDf(iniDf, null.ok = TRUE)
#> type <- match.arg(type)
#> .mv <- rxUdfUiMv()
#> if (!is.null(.mv)) {
#> .varsMv <- c(.mv$lhs, .mv$params, .mv$state)
#> .pre <- paste0(.var, num, rxIntToLetter(seq_len(power +
#> ifelse(intercept, 1L, 0L)) - 1L))
#> .pre <- vapply(.pre, function(v) {
#> if (v %in% .varsMv) {
#> paste0("rx.linMod.", v)
#> }
#> else {
#> v
#> }
#> }, character(1), USE.NAMES = FALSE)
#> }
#> else {
#> .pre <- paste0("rx.linMod.", .var, num, rxIntToLetter(seq_len(power +
#> ifelse(intercept, 1L, 0L)) - 1L))
#> }
#> if (!is.null(iniDf)) {
#> .theta <- iniDf[!is.na(iniDf$ntheta), , drop = FALSE]
#> if (length(.theta$ntheta) > 0L) {
#> .maxTheta <- max(.theta$ntheta)
#> .theta1 <- .theta[1, ]
#> }
#> else {
#> .maxTheta <- 0L
#> .theta1 <- .rxBlankIni("theta")
#> }
#> .theta1$lower <- -Inf
#> .theta1$upper <- Inf
#> .theta1$fix <- FALSE
#> .theta1$label <- NA_character_
#> .theta1$backTransform <- NA_character_
#> .theta1$condition <- NA_character_
#> .theta1$err <- NA_character_
#> .est <- rep(0, length(.pre))
#> if (data) {
#> .dat <- rxUdfUiData()
#> .wdv <- which(tolower(names(.dat)) == tolower(.dv))
#> if (length(.wdv) == 0L) {
#> warning(.dv, "not found in data, so no initial estimates will be set to zero")
#> }
#> else {
#> names(.dat)[.wdv] <- .dv
#> .model <- stats::lm(stats::as.formula(paste0(.dv,
#> " ~ stats::poly(", .var, ",", power, ")", ifelse(intercept,
#> "", "+0"))), data = rxUdfUiData())
#> .est <- coef(.model)
#> }
#> }
#> .cur <- c(list(.theta), lapply(seq_along(.pre), function(i) {
#> .cur <- .theta1
#> .cur$name <- .pre[i]
#> .cur$est <- .est[i]
#> .cur$ntheta <- .maxTheta + i
#> .cur
#> }))
#> .theta <- do.call(rbind, .cur)
#> .eta <- iniDf[is.na(iniDf$neta), , drop = FALSE]
#> .iniDf <- rbind(.theta, .eta)
#> }
#> else {
#> .iniDf <- NULL
#> }
#> .linMod <- paste(vapply(seq_along(.pre), function(i) {
#> if (intercept) {
#> if (i == 1)
#> return(.pre[i])
#> if (i == 2)
#> return(paste0(.pre[i], "*", .var))
#> paste0(.pre[i], "*", paste0(.var, "^", i - 1L))
#> }
#> else {
#> if (i == 1)
#> return(paste0(.pre[i], "*", .var))
#> paste0(.pre[i], "*", paste0(.var, "^", i))
#> }
#> }, character(1)), collapse = "+")
#> if (type == "replace") {
#> list(replace = .linMod, iniDf = .iniDf)
#> }
#> else if (type == "before") {
#> .replace <- paste0("rx.linMod.", .var, ".f", num)
#> list(before = paste0(.replace, " <- ", .linMod), replace = .replace,
#> iniDf = .iniDf)
#> }
#> else if (type == "after") {
#> .replace <- paste0("rx.linMod.", .var, ".f", num)
#> list(after = paste0(.replace, " <- ", .linMod), replace = "0",
#> iniDf = .iniDf)
#> }
#> }
#> <bytecode: 0x5555b7320e28>
#> <environment: namespace:rxode2>
# You can also print the s3 method that is used for this method
rxode2:::rxUdfUi.linMod
#> function (fun)
#> {
#> eval(fun)
#> }
#> <bytecode: 0x5555b7d59480>
#> <environment: namespace:rxode2>
#> attr(,"nargs")
#> [1] 2