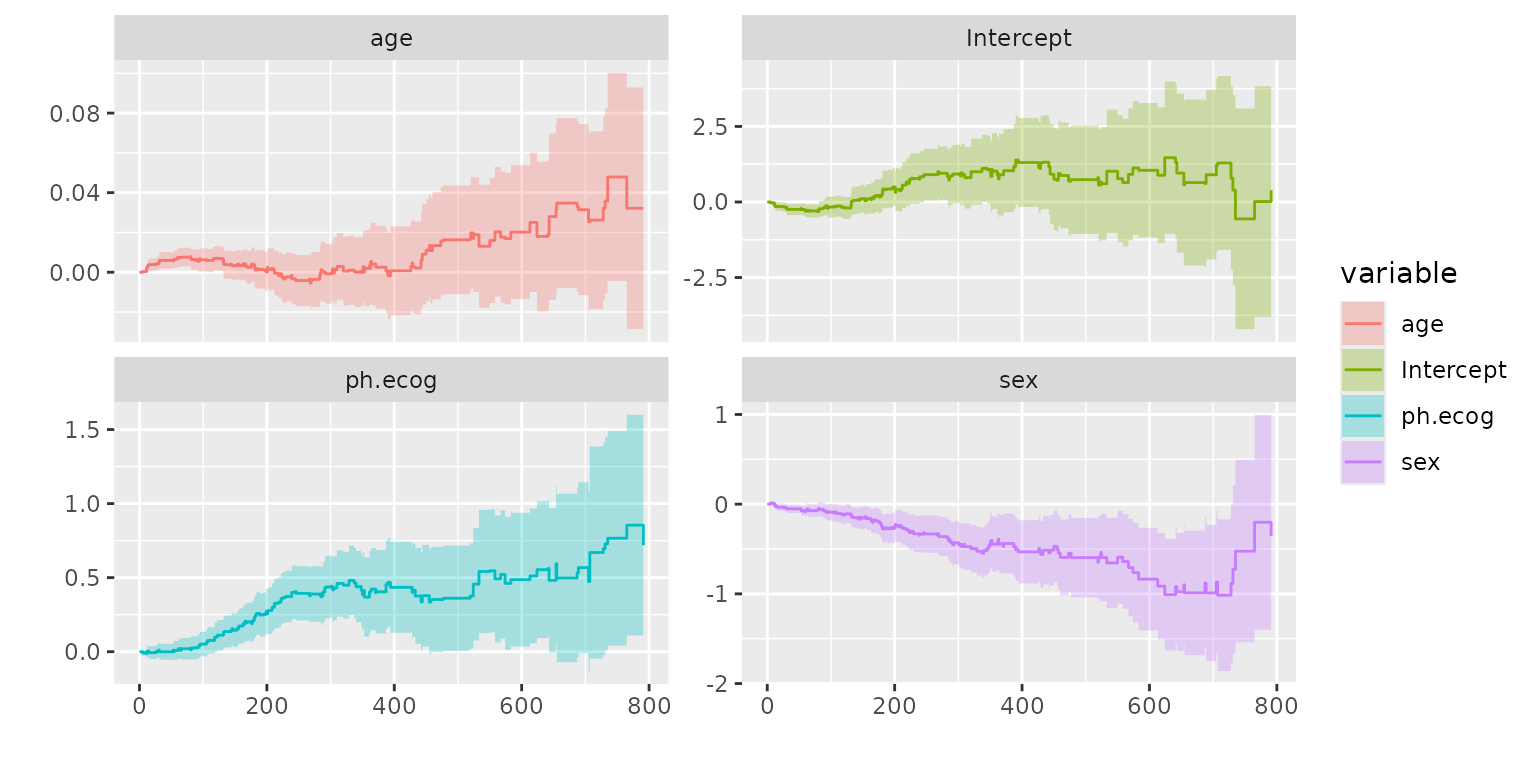
Source:
vignettes/plot_surv.Rmd
plot_surv.RmdThis document explains Survival Curves related plotting using ggplot2 and ggfortify.
Plotting with survival package
ggfortify let ggplot2 know how to draw survival curves. After loading ggfortify, you can use ggplot2::autoplot function for survfit objects.
library(ggfortify)
library(survival)
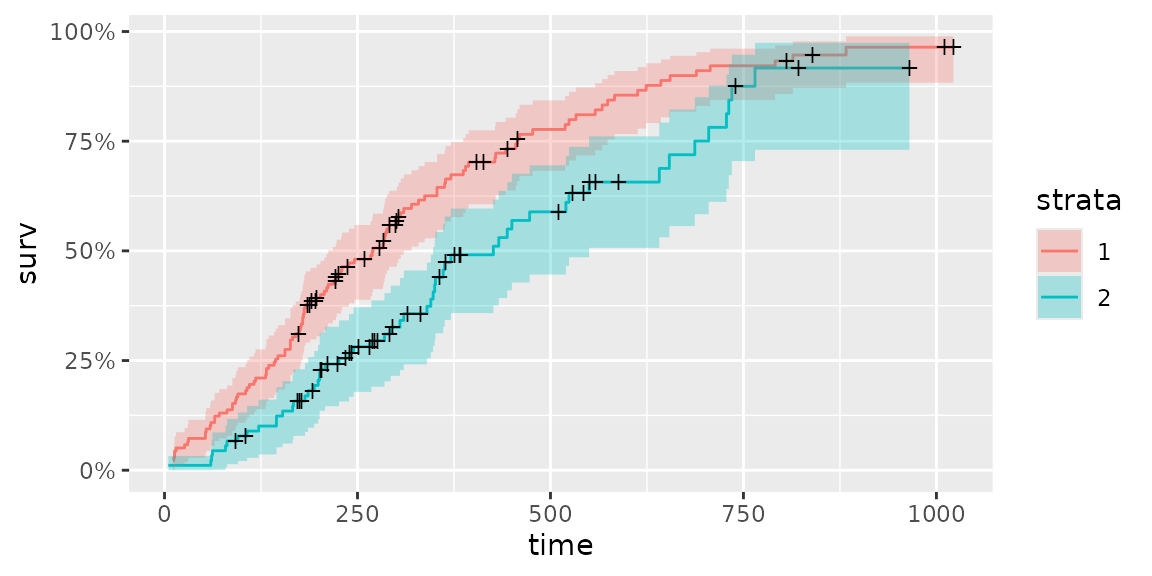
fit <- survfit(Surv(time, status) ~ sex, data = lung)
autoplot(fit)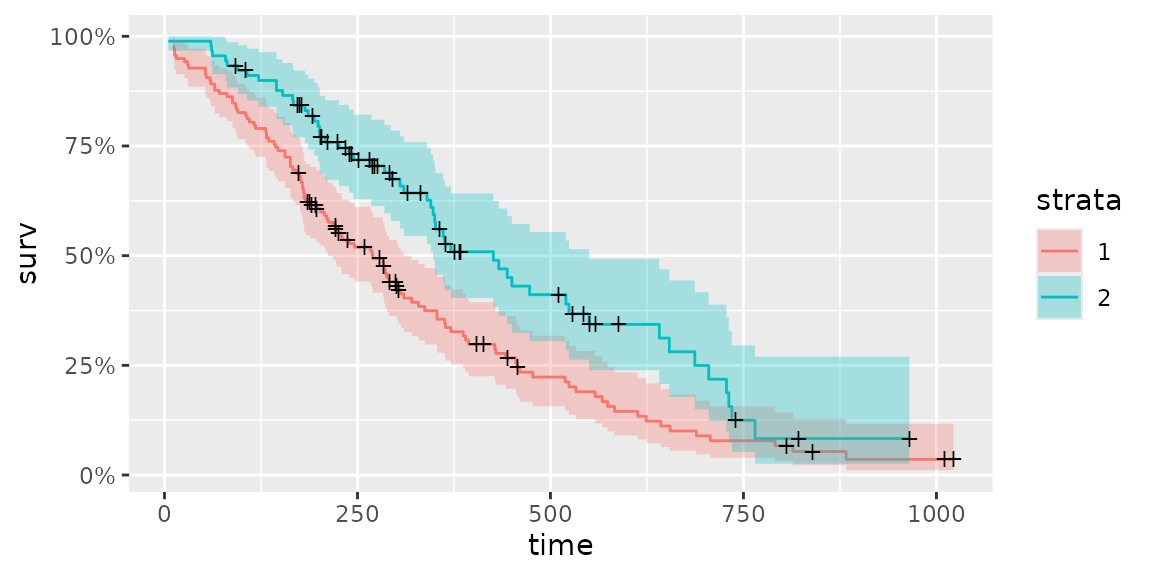
There are some options to change survival curve output. Use help(autoplot.survfit) (or help(autoplot.*) for any other objects) to check available options.
autoplot(fit, surv.linetype = 'dashed', conf.int = FALSE,
censor.shape = '*', censor.size = 5, facets = TRUE, ncol = 2)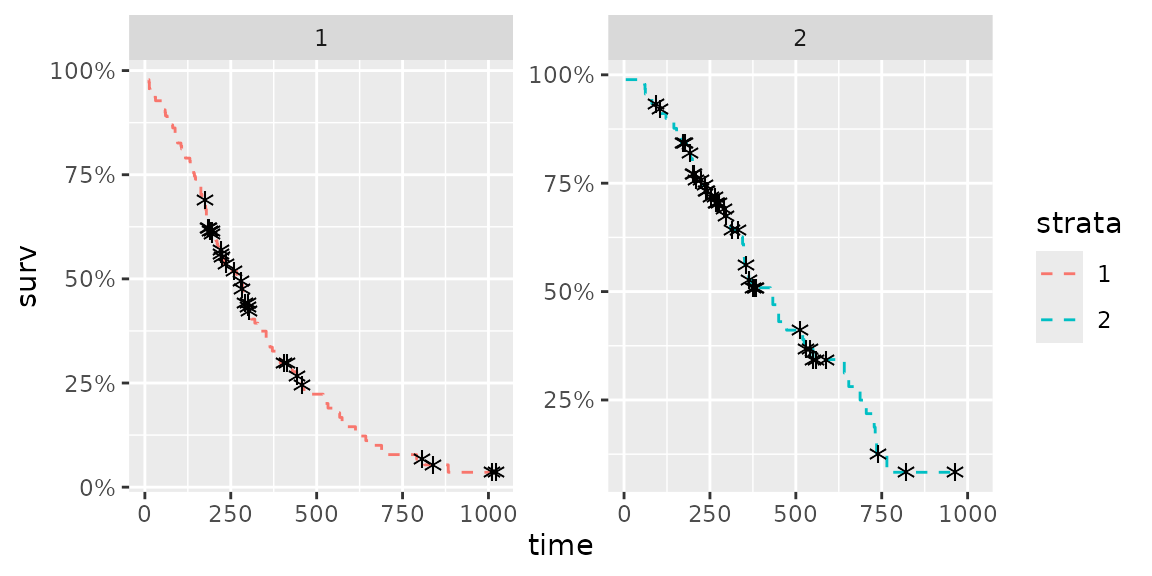
autoplot(survfit(Surv(time, status) ~ 1, data = lung), surv.colour = 'orange', censor.colour = 'red')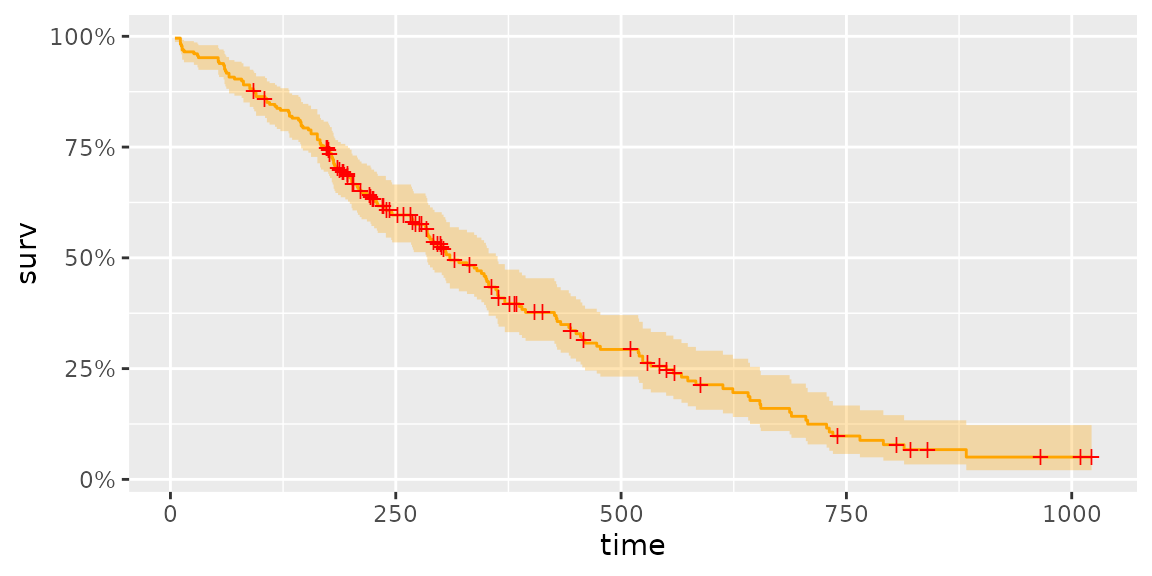
d.coxph <- survfit(coxph(Surv(time, status) ~ sex, data = lung))
autoplot(d.coxph, surv.linetype = 'dashed', surv.colour = 'blue',
conf.int.fill = 'dodgerblue3', conf.int.alpha = 0.5, censor = FALSE)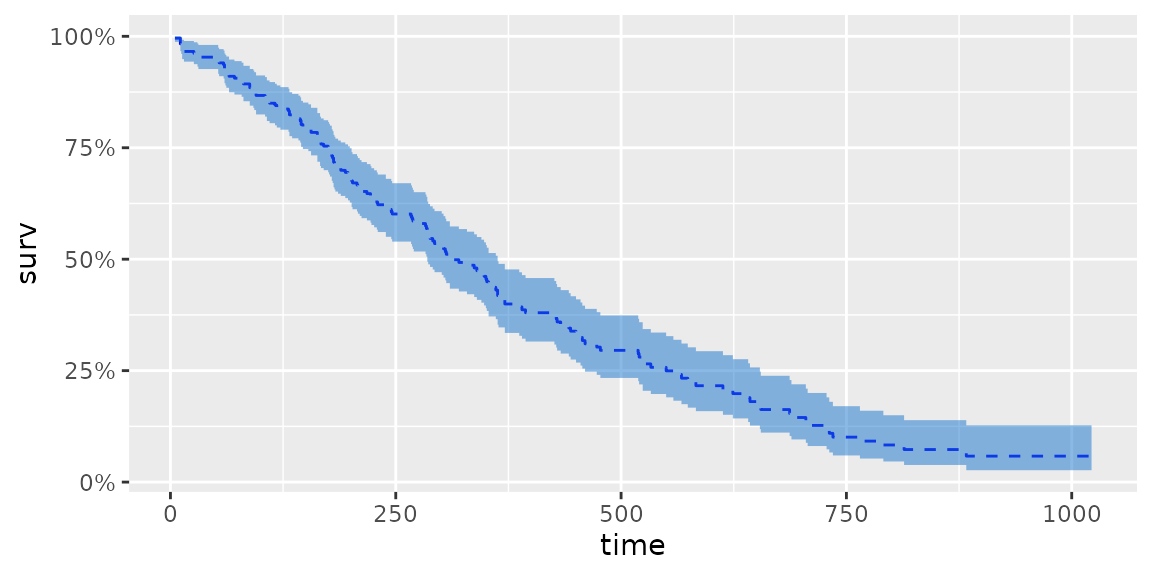
Also, you can use autoplot for survival::aareg instance.