Huberization – Bringing Outliers In
huberize.RdHuberization (named after Peter Huber's M-estimation algorithm for
location originally) replaces outlying values in a sample x by
their respective boundary: when \(x_j < c_1\) it is replaced by \(c_1\)
and when \(x_j > c_2\) it is replaced by \(c_2\). Consequently,
values inside the interval \([c_1, c_2]\) remain unchanged.
Here, \(c_j = M \pm c\cdot s\) where \(s := s(x)\) is
the robust scale estimate Qn(x) if that is positive,
and by default, \(M\) is the robust huber estimate of location
\(\mu\) (with tuning constant \(k\)).
In the degenerate case where Qn(x) == 0, trimmed means of
abs(x - M) are tried as scale estimate \(s\), with decreasing
trimming proportions specified by the decreasing trim vector.
Arguments
- x
numeric vector which is to be huberized.
- M
a number; defaulting to
huberM(x, k), the robust Huber M-estimator of location.- c
a positive number, the tuning constant for huberization of the sample
x.- trim
a decreasing vector of trimming proportions in \([0, 0.5]\), only used to trim the absolute deviations from
Min caseQn(x)is zero.- k
used if
Mis not specified as huberization centerM, and so, by default is taken as Huber's M-estimatehuberM(x, k).- warn0
logicalindicating if a warning should be signalled in caseQn(x)is zero and the trimmed means for all trimming proportionstrimare zero as well.- saveTrim
a
logicalindicating if the last triedtrim[j]value should be stored ifQn(x)was zero.
Details
In regular cases,
s = Qn(x)is positive and used to huberize values ofxoutside[M - c*s, M + c*s].In degenerate cases where
Qn(x) == 0, we search for an \(s > 0\) by trying the trimmed means := mean(abs(x-M), trim = trim[j])with less and less trimming (as the trimming proportionstrim[]must decrease). If even the last,trim[length(trim)], leads to \(s = 0\), a warning is printed whenwarn0is true.
Value
a numeric vector as x; in case Qn(x) was zero and
saveTrim is true, also containing the (last) trim
proportion used (to compute the scale \(s\)) as attribute "trim"
(see attr(), attributes).
Note
For the use in mc() and similar cases where mainly numerical
stabilization is necessary, a large c = 1e12 will lead to no
huberization, i.e., all y == x for y <- huberize(x, c)
for typical non-degenerate samples.
Examples
## For non-degenerate data and large c, nothing is huberized,
## as there are *no* really extreme outliers :
set.seed(101)
x <- rnorm(1000)
stopifnot(all.equal(x, huberize(x, c=100)))
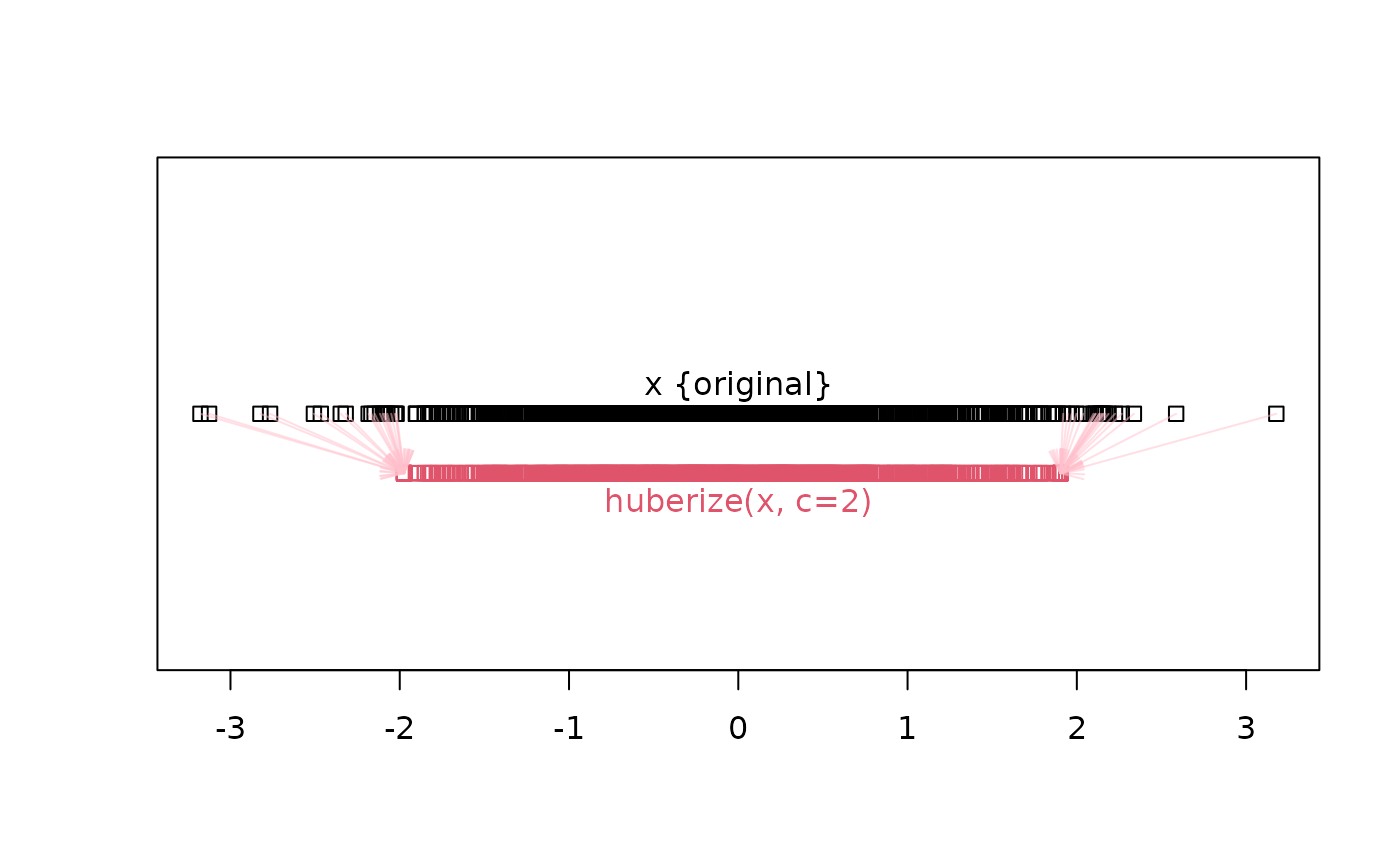
## OTOH, the "extremes" are shrunken towards the boundaries for smaller c:
xh <- huberize(x, c = 2)
table(x != xh)
#>
#> FALSE TRUE
#> 960 40
## 45 out of a 1000:
table(xh[x != xh])# 26 on the left boundary -2.098 and 19 on the right = 2.081
#>
#> -1.97404853359006 1.90241504050778
#> 20 20
## vizualization:
stripchart(x); text(0,1, "x {original}", pos=3); yh <- 0.9
stripchart(xh, at = yh, add=TRUE, col=2)
text(0, yh, "huberize(x, c=2)", col=2, pos=1)
arrows( x[x!=xh], 1,
xh[x!=xh], yh, length=1/8, col=adjustcolor("pink", 1/2))