Residuals of Robust Generalized Linear Model Fits
residuals.glmrob.RdCompute residuals of a fitted glmrob model, i.e., robust
generalized linear model fit.
Arguments
- object
an object of class
glmrob, typically the result of a call toglmrob.- type
the type of residuals which should be returned. The alternatives are:
"deviance"(default),"pearson","working","response", and"partial".- ...
further arguments passed to or from other methods.
Details
The references in glm define the types of residuals:
Davison & Snell is a good reference for the usages of each.
The partial residuals are a matrix of working residuals, with each column formed by omitting a term from the model.
The residuals (S3) method (see methods) for
glmrob models has been modeled to follow closely the
method for classical (non-robust) glm fitted models.
Possibly, see its documentation, i.e., residuals.glm, for
further details.
See also
glmrob for computing object, anova.glmrob;
the corresponding generic functions, summary.glmrob,
coef,
fitted,
residuals.
References
See those for the classical GLM's, glm.
Examples
### -------- Gamma family -- data from example(glm) ---
clotting <- data.frame(
u = c(5,10,15,20,30,40,60,80,100),
lot1 = c(118,58,42,35,27,25,21,19,18),
lot2 = c(69,35,26,21,18,16,13,12,12))
summary(cl <- glm (lot1 ~ log(u), data=clotting, family=Gamma))
#>
#> Call:
#> glm(formula = lot1 ~ log(u), family = Gamma, data = clotting)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -0.0165544 0.0009275 -17.85 4.28e-07 ***
#> log(u) 0.0153431 0.0004150 36.98 2.75e-09 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> (Dispersion parameter for Gamma family taken to be 0.002446059)
#>
#> Null deviance: 3.51283 on 8 degrees of freedom
#> Residual deviance: 0.01673 on 7 degrees of freedom
#> AIC: 37.99
#>
#> Number of Fisher Scoring iterations: 3
#>
summary(ro <- glmrob(lot1 ~ log(u), data=clotting, family=Gamma))
#>
#> Call: glmrob(formula = lot1 ~ log(u), family = Gamma, data = clotting)
#>
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) -0.0165260 0.0008369 -19.75 <2e-16 ***
#> log(u) 0.0153664 0.0003738 41.11 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#> Robustness weights w.r * w.x:
#> [1] 1.0000 0.6208 1.0000 1.0000 1.0000 1.0000 1.0000 1.0000 1.0000
#>
#> Number of observations: 9
#> Fitted by method ‘Mqle’ (in 3 iterations)
#>
#> (Dispersion parameter for Gamma family taken to be 0.001869399)
#>
#> No deviance values available
#> Algorithmic parameters:
#> acc tcc
#> 0.0001 1.3450
#> maxit
#> 50
#> test.acc
#> "coef"
#>
clotM5.high <- within(clotting, { lot1[5] <- 60 })
cl5.high <- glm (lot1 ~ log(u), data=clotM5.high, family=Gamma)
ro5.high <- glmrob(lot1 ~ log(u), data=clotM5.high, family=Gamma)
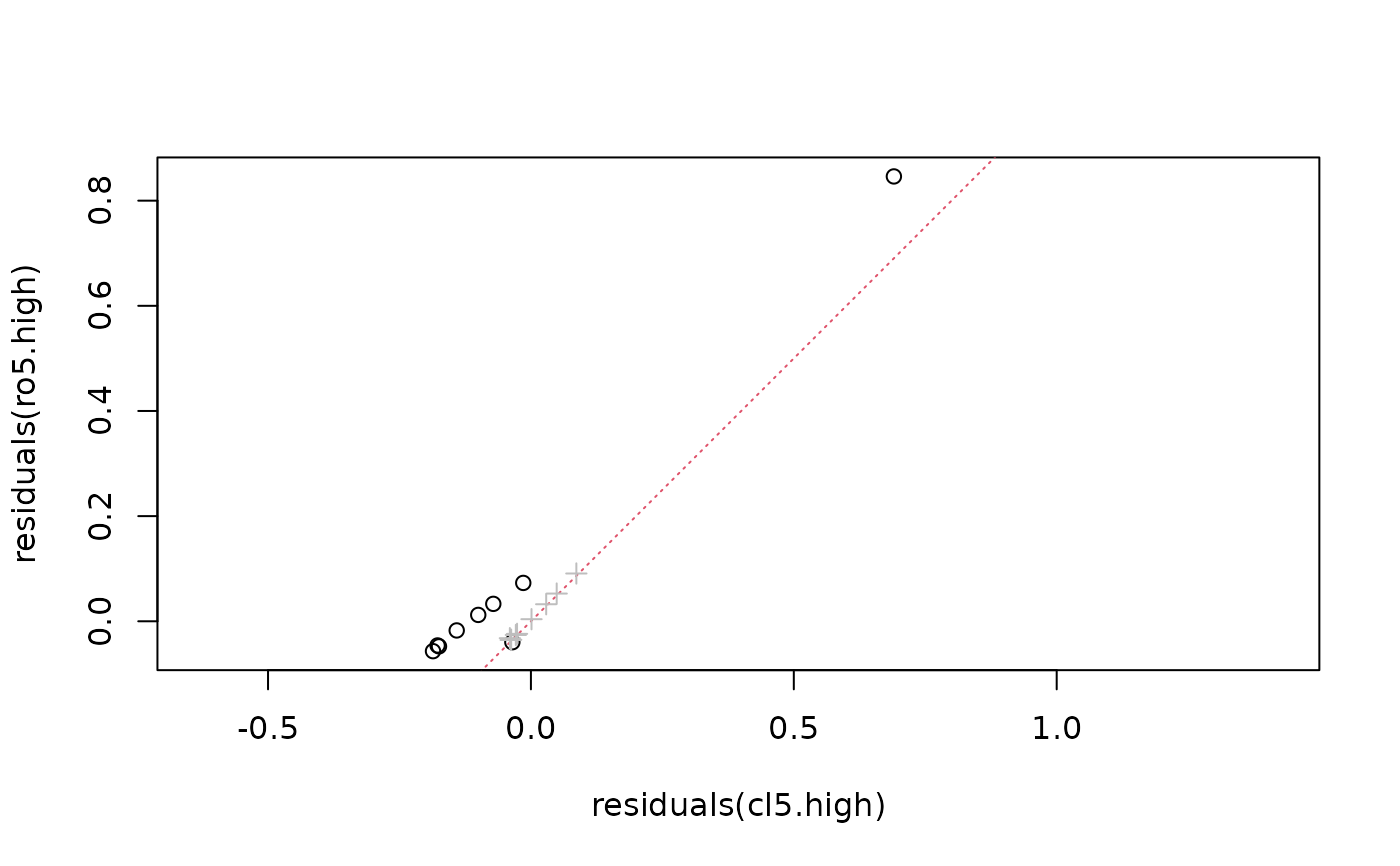
rr <- range(residuals(ro), residuals(cl), residuals(ro5.high))
plot(residuals(ro5.high) ~ residuals(cl5.high), xlim = rr, ylim = rr, asp = 1)
abline(0,1, col=2, lty=3)
points(residuals(ro) ~ residuals(cl), col = "gray", pch=3)
 ## Show all kinds of residuals:
r.types <- c("deviance", "pearson", "working", "response")
sapply(r.types, residuals, object = ro5.high)
#> deviance pearson working response
#> 1 -0.03981550 -0.03928883 -0.03928883 -4.8256777
#> 2 0.07296761 0.07475306 0.07475306 4.0341149
#> 3 0.03313416 0.03350112 0.03350112 1.3614374
#> 4 0.01211012 0.01215906 0.01215906 0.4204547
#> 5 0.84608494 1.09974648 1.09974648 31.4251217
#> 6 -0.01741507 -0.01731412 -0.01731412 -0.4404795
#> 7 -0.04768288 -0.04692803 -0.04692803 -1.0340127
#> 8 -0.05684730 -0.05577523 -0.05577523 -1.1223275
#> 9 -0.04597419 -0.04527236 -0.04527236 -0.8535445
## Show all kinds of residuals:
r.types <- c("deviance", "pearson", "working", "response")
sapply(r.types, residuals, object = ro5.high)
#> deviance pearson working response
#> 1 -0.03981550 -0.03928883 -0.03928883 -4.8256777
#> 2 0.07296761 0.07475306 0.07475306 4.0341149
#> 3 0.03313416 0.03350112 0.03350112 1.3614374
#> 4 0.01211012 0.01215906 0.01215906 0.4204547
#> 5 0.84608494 1.09974648 1.09974648 31.4251217
#> 6 -0.01741507 -0.01731412 -0.01731412 -0.4404795
#> 7 -0.04768288 -0.04692803 -0.04692803 -1.0340127
#> 8 -0.05684730 -0.05577523 -0.05577523 -1.1223275
#> 9 -0.04597419 -0.04527236 -0.04527236 -0.8535445